Researchers identify a distinct new CRISPR editing platform
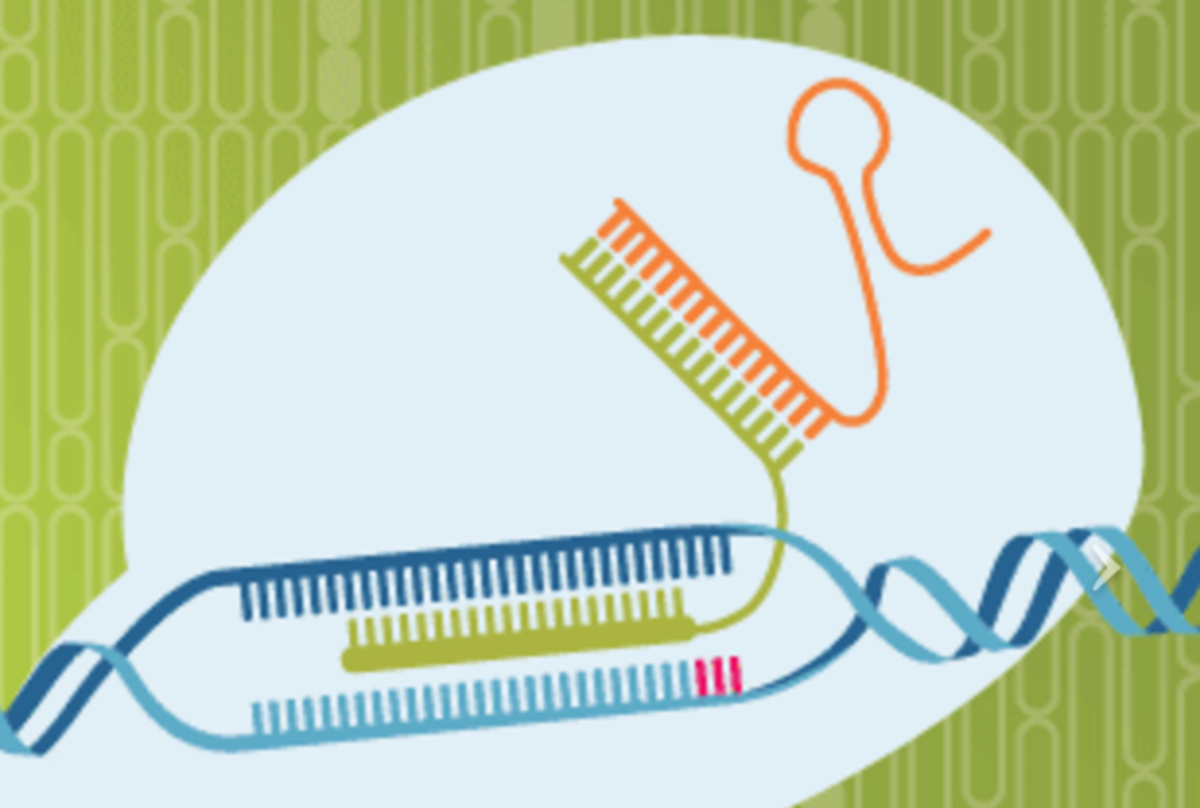
The novel CasX variants were first described by Doudna and colleagues in 2016. In their recent Nature publication, they now demonstrate that CasX is indeed an RNA-guided DNA endonuclease that generates a staggered double-stranded break in DNA at sequences complementary to a 20-nucleotide segment of its guide RNA, and that it can be applied to genome editing in human cells.
Interestingly, CasX contains several domains distinct from those found in Cas9 and Cas12a, as well as novel RNA and protein folds. The authors therefore establish CasX as a novel, third CRISPR-Cas platform. They also note that the small size of CasX (<1,000 amino acids), its DNA cleavage characteristics, and its derivation from non-pathogenic microorganisms may offer important advantages over existing CRISPR–Cas platforms.
CasX uses a 20-nucleotide guide RNA segment and cleaves adjacent to a TTCN PAM, generating products with staggered ends about 10 nucleotides in length. Furthermore, it is capable of inducing cleavage and gene editing in ways that are different than Cas9 and Cas12a. The authors write "It is clear that CasX possesses domains analogous to other CRISPR-Cas proteins as well as completely novel domains, as anticipated by its complete sequence dissimilarity from other CRISPR proteins. A prominent guide RNA scaffold for CasX structural modelling shows that the guide RNA accounts for about 26 percent of the mass in the CasX-sgRNA binary complex, which is a value that is greater than those observed for other type II or type V CRISPR–Cas effector complexes (about 8 percent in LbCas12a, about 20 percent in Alicyclobacillus acidoterrestris Cas12b, and about 16 percent in SpCas9."
The compact size of CasX together with its dominant RNA content could enable its development into a third platform for RNA-programmed genome editing. The nuclease may also be used for therapeutic delivery and may ultimately be safer than existing genome-editing technologies.