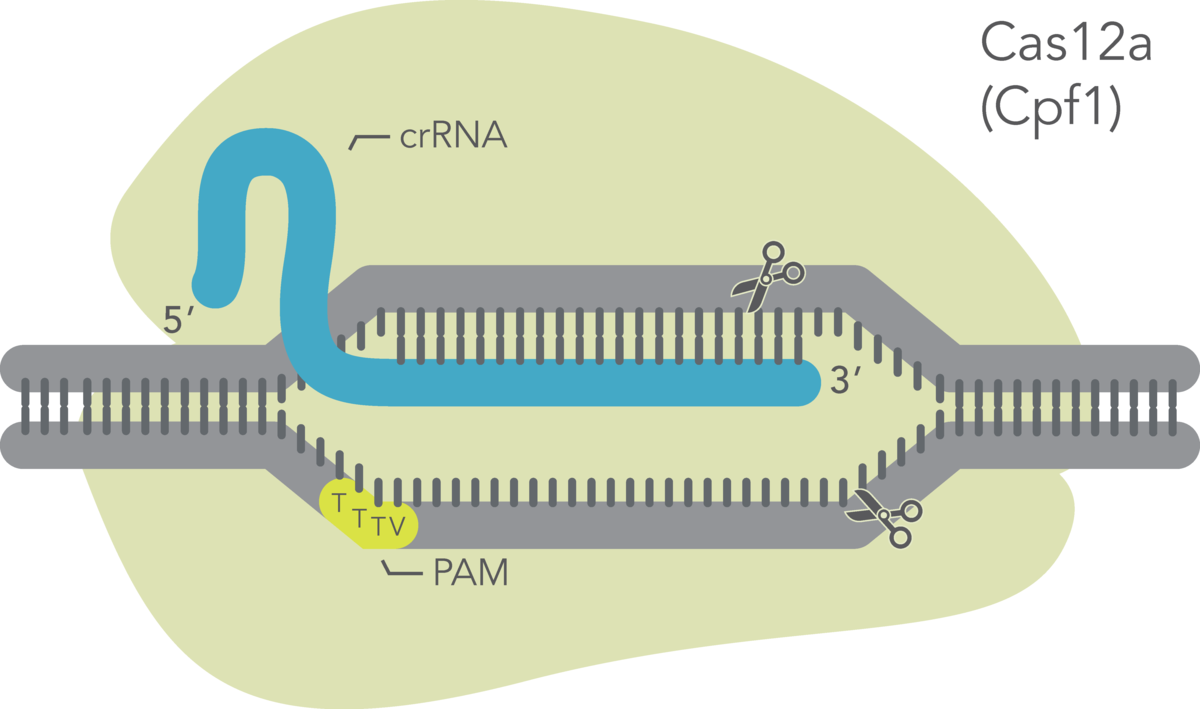
CRISPR-Cas12a genome editing method uses the Cas12a endonuclease to generate double-stranded breaks that contain a staggered 5′ overhang. Cas12a requires only a single CRISPR RNA (crRNA) to specify the DNA target sequence. After cleavage, DNA is then repaired by non-homologous end-joining (NHEJ) or homology-directed recombination (HDR), resulting in a modified sequence. Alt-R™ CRISPR-Cas12a reagents from our supplier Integrated DNA Technologies (IDT) provide essential, optimized tools needed to use this pathway for genome editing research. The system enables targeting of additional genomic sites that are not available to the CRISPR-Cas9 System.
Overview of Alt-R™ CRISPR-Cas12a System
| Required components | |
|---|---|
| Alt-R CRISPR-Cas12a (Cpf1) crRNA | Target-specific RNA oligo, custom synthesized based on your sequence |
Alt-R A.s. or L.b. Cas12a (Cpf1) Ultra (recommended) |
|
| Alt-R Cas12a (Cpf1) Electroporation Enhancer | Cas12a-specific carrier DNA highly recommended for efficient electroporation of the RNP |
| Related reagents and kits | |
|---|---|
| Alt-R HDR Enhancer V2 | For improved rates of homology-directed repair |
| Cas12a positive controls | Order as custom crRNAs or oligos
|
| Alt-R Genome Editing Detection Kit | For mutation detection and estimating editing efficiency |
Guide RNA for the Cas12a system
The Alt-R CRISPR-Cas12a (Cpf1) crRNA is a single, 40–44 base, guide RNA, comprising a 20 base constant region (loop domain) and a 20–24 base target-specific region (protospacer domain). IDT typically recommends a 21 base protospacer domain for optimal activity. All Alt-R CRISPR-Cas12a (Cpf1) crRNAs are synthesized with proprietary chemical modifications, which protect the crRNA from degradation by cellular RNases and further improve on-target editing performance. For crRNA design, identify locations in your target region with the protospacer adjacent motif (PAM) sequence, TTTV, where V is A, C, or G. If you are using Alt-R Cas12a Ultra, a TTTT PAM sequence may also work but may not be as potent. Your Alt-R CRISPR-Cas12a (Cpf1) crRNA will bind to the DNA strand opposite to the PAM sequence. Do not include the PAM sequence in your crRNA design. An example of a correct crRNA sequence is shown in the following figure. Common incorrect design examples are shown as crossed-out gray text:
Order crRNA for the Cas12a system on the website of our supplier IDT. Because the crRNA recognizes and binds 21 bases on the DNA strand opposite from the TTTV sequence of the PAM site, order your crRNA by entering the 20–24 bases downstream of the PAM site, in the forward orientation as shown. Enter only DNA bases into the order entry tool. If you are pasting your CRISPR target site from an online design tool, make sure you verify the correct strand orientation. After you enter your 20–24 base target sequence, 20 additional bases and the necessary modifications will automatically be added by the order entry system for a total of 40–44 RNA bases.
Cas12a nucleases
LubioScience offers Cas12a nucleases from two different species, Acidaminococcus sp. BV3L6 (A.s.) and Lachnospiraceae bacterium ND2006 (L.b.). In general, we recommend using Alt-R Cas12a (Cpf1) Ultra nucleases, which have higher on-target potency than their wild-type A.s. or L.b. Cas12a nuclease counterparts. Cas12a Ultra reaches or even exceeds the performance of Cas9. The Ultra mutants recognize many TTTT PAM sites in addition to TTTV motifs, which increases their target range for genome editing studies. In addition, the Alt-R L.b. Cas12a (Cpf1) Ultra mutant has been shown to have increased temperature tolerance, making it an ideal solution for systems that require lower culture temperatures, such as those needed for plant cultures. The wild-type Alt-R A.s. Cas12a V3 enzyme is also available for legacy use.
Alt-R A.s. Cas12a (Cpf1) nuclease
Alt-R A.s. Cas12a (Cpf1) Ultra and V3 nucleases are high-purity, recombinant Acidaminococcus sp. Cas12a. The enzymes include nuclear localization sequences and C-terminal 6-His tags. The Cas12a enzyme should be combined with a crRNA to produce a functional, target-specific editing complex. For the best editing, combine Alt-R A.s. Cas12a Ultra or V3 enzyme with optimized Alt-R CRISPR-Cas12a crRNA in equimolar amounts. The A.s. Cas12a PAM sequence is TTTV, which permits targeting of DNA sequences in AT-rich regions of the genome. The Alt-R A.s. Cas12a Ultra enzyme has increased recognition of the TTTT PAM sequence.
Alt-R L.b. Cas12a (Cpf1) Ultra nuclease
Alt-R L.b. Cas12a (Cpf1) Ultra nuclease is high-purity, recombinant Lachnospiraceae bacterium ND2006 nuclease, purified from an E. coli strain expressing Cas12a with proprietary mutations to improve both on-target performance and temperature tolerance. The enzyme includes nuclear localization sequences and C-terminal 6-His tags. The Cas12a enzyme should be combined with a crRNA to produce a functional, target-specific editing complex. For the best editing, combine Alt-R L.b. Cas12a Ultra enzyme with optimized Alt-R CRISPR-Cas12a crRNA in equimolar amounts. L.b. Cas12a (Cpf1) Ultra mutant has been shown to have increased temperature tolerance for systems that require lower culture temperatures.
Cas12a controls
As you plan your experiment, we recommend including positive and negative controls, so your downstream analysis will be as informative as possible. Some of the most important controls are gRNA sequences. A positive control should lead to cutting of a known target site. A negative control should not specifically cut genome sequences. Other negative controls can include samples with only vehicle, Cas protein, gRNA, or cells, as well as any other negative controls that are appropriate in your particular experiment.
Positive and negative control gRNA target sequences for Cas12a for human, rat, and mouse genomes are listed on IDT's website. You can order them there. Furthermore, IDT offers PCR primers (Alt-R HPRT PCR Primer Mixes for human or mouse) for use with their Alt-R Genome Editing Detection Kit to identify editing or estimate editing efficiency in samples transfected with the positive control crRNAs.
Additional resources
For more detailed information about gene editing using CRISPR-Cas12a and CRISPR gene editing in general, download IDT's CRISPR basics handbook. Further insights into the products can also be found on IDT's website.
Download handbook Go to IDT website Further reading and links
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend for these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.