Introduction
After delivery of the editing complex, you normally have a heterogeneous cell population consisting of unedited and edited cells. Edited cells may either be heterozygous (one allele edited) or homozygous (both alleles edited).
Depending on the goal of your experiment and your downstream analysis, you may either continue with a heterogeneous cell population or you may have to pick, analyse and expand individual edited cells.
Below you will find a number of assays, kits, reagents, and instructions which are useful for the analysis of your edited cell population as well as for cell line storage.
Isolation of edited cells
To isolate cells with your specific gene edit, you can create a monoclonal cell culture. There are different ways to clone the cells manually:
- Grow cells in a large plate and isolate them with glass cloning rings. That is, the transfected cells are plated at a very low concentration so that they form colonies in little patches. Then, glass cloning rings, dipped in sterile vacuum grease, can be placed around the colonies to keep them separate.
- Without using cloning rings, use a pipette tip to lift individual colonies from a large plate into the wells of multiwell plates.
- (Recommended) Dilute transfected cells and systematically transfer them into 96-well plates with less than 1 cell per well. Then, grow the cells until you see colonies in some of the wells. The reason this is frequently the best method is because there is less possibility of cross-contamination between clones and less complexity in pipetting than the other methods. Our supplier Integrated DNA Technologies (IDT) provides a detailed protocol for this technique with a description of the pros and cons.
DNA isolation from edited cells
DNA isolation using QuickExtract™
A simple, rapid extraction of PCR-ready DNA for screening your edited cells.
- 8-minute extraction protocol for most sample types
- No centrifugation steps or spin columns
- DNA extraction requires only heat treatment to lyse the cellular or tissue material, release the DNA, and degrade compounds inhibitory to amplification.
- Automation-friendly: a simple protocol that integrates easily into automated workflows
- Safe: uses only non-toxic reagents
- Recommended for rapid, easy sample prep for CRISPR mutation detection assays
| Cat-No. | Item | Size | Price (CHF) |
|---|---|---|---|
| QE09050 | QuickExtract DNA Extraction Solution 1.0, 50 mL | 50 ml | 523.00 |
DNA isolation kits
We offer a range of genomic DNA isolation kits from Zymo Research that are suitable for extracting high molecular weight DNA from a wide variety of sample types. These kits yield high-quality DNA that is ideal for use in any sensitive downstream applications such as DNA sequencing.
Cas9 ELISA Assay

The assay can be used to monitor S. pyogenes Cas9 in cell culture lysate and is ideal for screening experimental samples utilizing the CRISPR/Cas9 gene editing system.
Assay principle: Microtitration wells coated with murine anti-Cas9 capture antibody are exposed to test specimens, which may contain Cas9 reactive determinants. The Cas9 antigen in the specimen is specifically captured onto the immobilized antibody during specimen incubation. The captured antigen is then reacted with a biotinylated mouse monoclonal Cas9 detection antibody. Subsequently, Streptavidin-HRP conjugate is added. Following a wash cycle, specifically bound enzyme conjugate is detected by reaction with the Substrate Solution, tetramethylbenzidine (TMB). The assay is measured spectrophotometrically to indicate the level of Cas9 reactive determinants present in a sample.
Alt-R™ Genome Editing Detection Kit
The T7 endonuclease I (T7EI) mismatch cleavage assay is used in the Alt-R Genome Editing Detection Kit. This assay is useful for a quick, qualitative assessment of genome editing efficiency in CRISPR-treated cells. T7EI is an enzyme that cleaves double-stranded DNA with small mismatches between the two strands, so-called heteroduplexes. The following picture illustrates the principle of the assay:
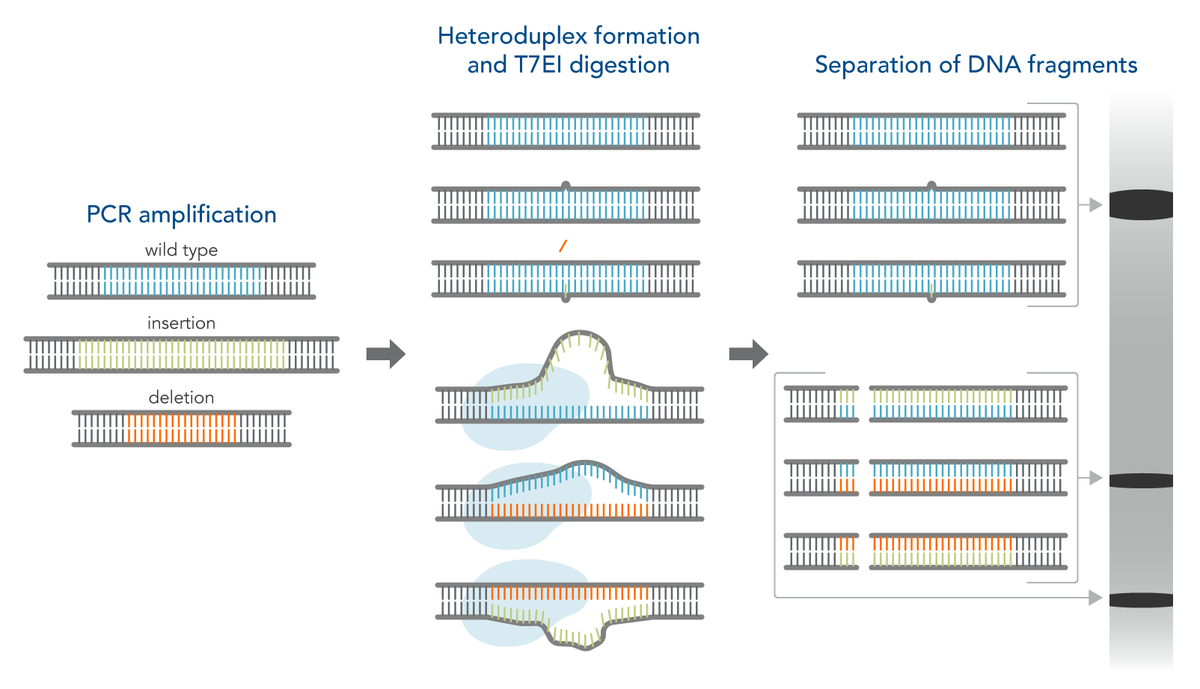
Design the PCR primers so that they amplify both the target sequence and flanking sequences on each side of the target. Researchers at IDT recommend amplifying a region of 600–1000 bp with at least 100 bp at each side of the cut site. The cut site should be off-center so that the two digestion products have different sizes that are distinguishable on a gel. To execute the T7EI assay, we recommend isolating DNA 48 hours after addition of the RNP and amplifying the target locus using PCR. The PCR products are melted at 95°C and slowly cooled, creating a mixture of homoduplexes (between complementary wildtype sequences) and heteroduplexes (between strands of DNA from PCR products that contain wildtype or CRISPR-edited sequences). The duplexes can then be digested with T7EI endonuclease to determine total editing.
Advantages of the Alt-R Genome Editing Detection Kit at a glance:
- Straightforward: Use standard molecular biology techniques
- Fast: Does not require purification of PCR products before analysis
- Consistent: Analyze easy-to-read electrophoresis results
- Quantifiable: Gel band intensities relate to genome editing efficiency
- Versatile: Convenient for a single sample or high throughput analysis in plate format
rhAmpSeq™ targeted sequencing for analysis of on- and off-target editing
Nominated hotspot sites for off-target effects can be verified with a targeted NGS library prep method, such as the IDT rhAmpSeq amplicon sequencing system. The rhAmpSeq system utilizes primers that reduce non-specific amplification for characterizing editing events by amplifying the targeted site for sequencing after you have performed genome editing. You can generate amplicons of the targeted site and of the nominated hotspots in a single reaction, and then you use NGS to sequence the amplicons.
The rhAmpSeq rapid library preparation protocol allows amplification of up to 5000 targets in a single PCR sample. The method therefore enables simultaneous testing of multiple editing sites, both on- and off-target. The rhAmpSeq system enables verification of edits with reduced off-target amplification and can be used in conjunction with Illumina instruments. The resulting data can be analyzed using the rhAmpSeq CRISPR Analysis Tool.
Learn more about the rhAmpSeq CRISPR Analysis System Order on IDT website
Storage of edited cell pools and clonal lines
Store your valuable cell pools and clonal cell lines using Bambanker™ in order to ensure maximum viability during long-term storage. Its innovative formulation simplifies the freezing process and greatly increases cell viability. With Bambanker, the viability of cells during freeze/thaw cycles is greatly enhanced, even for fragile cells such as embryonic stem cells.



Additional resources
For more detailed information about gene editing using CRISPR-Cas, download IDT's CRISPR basics handbook. Further insights into IDT's products for CRISPR genome engineering can also be found on their website.
Download handbook Go to IDT website Further reading and links
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend for these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.

