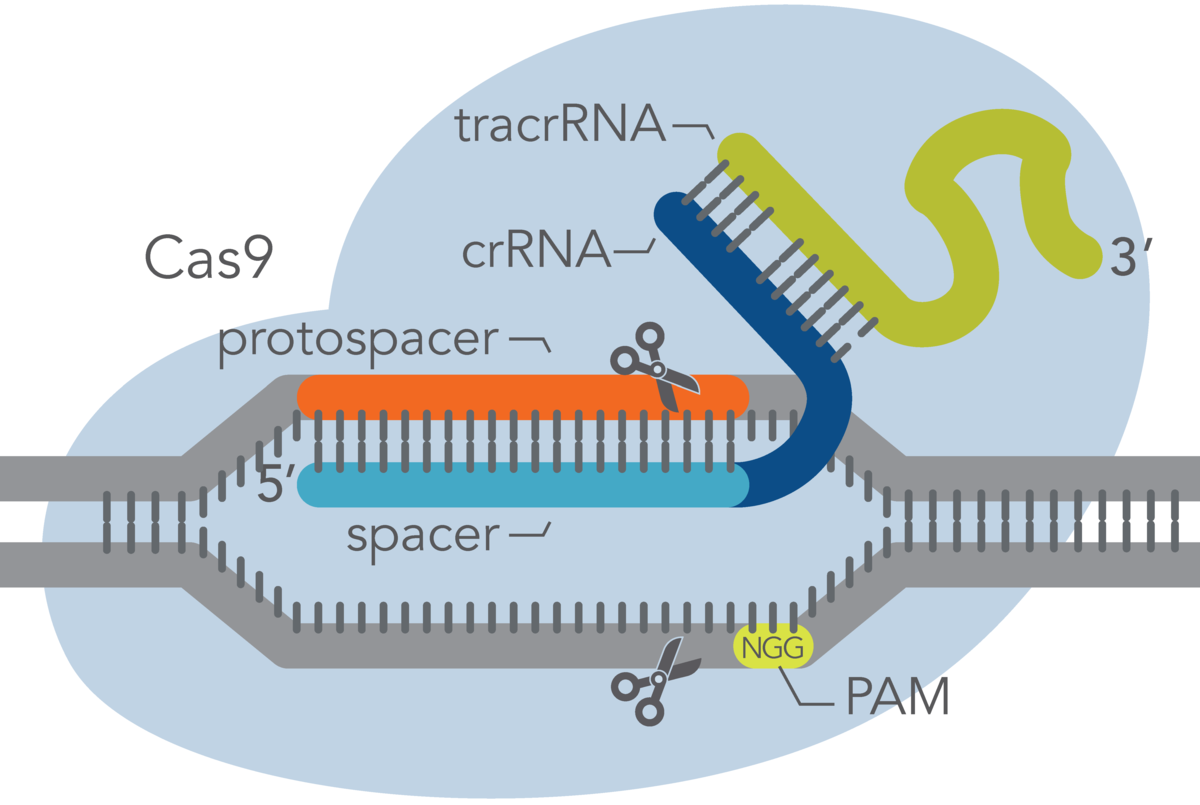
At the most basic level, CRISPR genome editing requires 2 components: a Cas enzyme and a guide RNA. LubioScience offers a broad range of products to support the success of your CRISPR-Cas9 experiment. Learn more about the Alt-R™ CRISPR-Cas9 System from our trusted supplier Integrated DNA Technologies (IDT). The product portfolio includes optimized guide RNAs, a gRNA design tool, various Cas9 enzymes suitable for your specific application, and control reagents.
Overview of Alt-R™ CRISPR-Cas9 System
| Ribonucleoprotein components | |
|---|---|
Option 1: Alt-R CRISPR-Cas9 crRNA:tracrRNA | Alt-R CRISPR-Cas9 crRNA
|
Alt-R CRISPR-Cas9 tracrRNA
| |
| Option 2: Alt-R CRISPR-Cas9 sgRNA |
|
| Alt-R S.p. Cas9 Nuclease/Nickase |
|
| Additional reagents and kits | |
|---|---|
| Alt-R CRISPR-Cas9 Control Kits |
|
| Alt-R CRISPR-Cas9 Electroporation Enhancer | For primary and difficult-to-transfect cells |
| Alt-R HDR Enhancer V2 | For improved rates of homology-directed repair |
| Alt-R Genome Editing Detection Kit | For mutation identification and estimating editing efficiency |
Guide RNA formats
2-part RNA system vs single guide RNA
All of IDT's Alt-R CRISPR RNA molecules contain chemical modifications, which protect from cellular endogenous nuclease activity. One initial decision is to decide whether to use a 2-part RNA system or a single guide RNA (sgRNA) system. For Cas9 CRISPR experiments, sometimes there are advantages to splitting the guide RNA into a tracrRNA and a crRNA molecule (2-part system), as found in nature. One obvious advantage is that tracrRNA is always the same across all experiments. Since the crRNA is short, it is relatively inexpensive. If you are targeting many sequences, you can simply get one large order of tracrRNA and use it with any of your crRNA sequences, saving cost compared to sgRNA. When using the 2-part guide RNA system in high nuclease environments, a more modified version of the guide RNA may be required. In this case, consider using one of our extra-modified gRNAs: the Alt-R 2-part XT guide RNA or Alt-R sgRNA, which both have high stability. The table below gives an overview of the different types of guide RNAs.
If you need help choosing which type of guide RNA is best for your Cas9 experiment, the following article provides more discussion and guidance about which gRNA format to use (quick tip: it usually does not make a difference).
Advantages of chemically synthesized and modified gRNA
Using one of IDT's synthetic gRNA options has several advantages over expressing the gRNA, including improved editing efficiency and reduced labor intensity. Preformed guide RNA also does not continue to be expressed over a long period of time like plasmid-encoded guide RNA. By minimizing how long the gRNAs spend in the target cells, synthetic options help decrease the chance of toxicity. Despite that, chemical modifications also increase the amount of time the gRNA is in the cells. The goal is to use the optimum time so that the gRNA is in the cells long enough to be effective, but not so long that the gRNA causes toxicity or increases off-target effects. IDT provides chemically synthesized guide RNAs with optimized formats.
All of the IDT Alt-R CRISPR RNA molecules contain chemical modifications, which protect from cellular endogenous nuclease activity. In addition, if you are still worried that high levels of endogenous, cellular nucleases might destroy the crRNA in your experiment, you can choose to use IDT's XT modification on crRNA. XT crRNA contains additional chemical modifications, making it longer-lived, nuclease resistant, safe for your cells, and still just as functional for CRISPR as regular crRNA. The standard Alt-R crRNA is end-blocked, whereas Alt-R crRNA-XT has both end-blocking and internal chemical modifications. Similarly, the Alt-R tracrRNA is heavily modified.
Guide RNA design considerations for Cas9
On- and off-target activity
The goal when designing a guide RNA is achieving the highest possible on-target activity of your CRISPR gene editing complex, while minimizing off-target activity. Off-target activity causes unwanted phenotypes, potentially including cell death.
PAM sequence
For Cas9 guide RNA designs, the target sequence must be next to a PAM sequence, NGG, where N is any base. It is important that you do not include the PAM sequence in the actual guide RNA design. The guide RNA recognizes and binds to 20 nucleotides on the DNA strand opposite from the NGG PAM site. The “N” of the NGG is immediately adjacent to the most 3’ base of the non-targeted strand side of the protospacer.
Length considerations
When you are using 2-part guide RNA, there are crRNA and tracrRNA components:
- crRNA: The crRNA is composed of a target-specific spacer region and another domain that hybridizes to the tracrRNA. The target-specific spacer sequence is 20 nucleotides long; if it is shorter than this, the on-target activity will be negatively impacted. IDT researchers have found that the optimal total length of the crRNA (the target-specific spacer region plus the domain that hybridizes to tracrRNA) is 36 nucleotides.
- tracrRNA: Part of the tracrRNA molecule hybridizes to the crRNA, and another part of it binds to Cas9. IDT scientists have found that the optimal, total length for tracrRNA is 67 nucleotides. This is shorter than the tracrRNA found in nature, but IDT scientists have found that when using 2-part guide RNA, shortening the tracrRNA to 67 nucleotides increases on-target activity.
When using sgRNA, the crRNA and tracrRNA are all part of one longer RNA molecule joined by a hairpin-like loop. Cas9 sgRNAs for genome-editing purposes are typically 100 nucleotides in length.
IDT guide RNA design tool
A good design tool uses optimized rules for maximizing on-target editing, while checking for (and minimizing) off-target effects, based both on experimental data and bioinformatics. IDT offers a free CRISPR guide RNA design tool that has three modes:
- Search for predesigned gRNA: Select from predesigned Alt-R CRISPR-Cas9 guide RNAs (gRNAs, such as crRNA and sgRNA) targeting human, mouse, rat, zebrafish, or C. elegans gene targets.
- Design custom gRNA: Generate CRISPR-Cas9 guide RNAs (gRNAs, such as crRNA and sgRNA) targeting any sequence from any species. Currently, analysis of off-target effects against human, mouse, rat, zebrafish, or C. elegans genes is available.
- CRISPR-Cas9 gRNA checker: Assess on- and off-targeting potential of protospacer designs of your own or from publications before ordering guide RNAs (gRNAs, such as crRNA and sgRNA) that are synthesized using Alt-R gRNA modifications.
For HDR experiment designs, please see the following HDR design tool.
Choosing a Cas9 enzyme
Cas9, the best-known and most popular CRISPR-associated nuclease, can target any ~20-nucleotide DNA sequence with an adjacent NGG PAM sequence (where N is any base). There are 3 main types of Alt-R S.p. Cas9 enzymes available from IDT: wild-type (WT), HiFi, and nickase variants. Please refer to the provided table for a direct comparison of the three.
| Alt-R S.p. Wild-type Cas9 Nuclease | Alt-R S.p. HiFi Cas9 Nuclease | Alt-R S.p. Cas9 D10A Nickase | Alt-R S.p. Cas9 H840A Nickase | |
|---|---|---|---|---|
| Description | Wild-type Cas9 with high genome editing potency that is simple to use and economical | Cas9 variant that effectively reduces off-target effects and preserves high on-target activity | Cas9 variant with a mutation in the RuvC domain that disables cleavage of the non-target strand | Cas9 variant with a mutation in the HNH domain that disables cleavage of the target strand |
| DNA cleavage | Both strands | Both strands | Target strand | Non-target strand |
| Suggested use | First choice for most CRISPR genome editing projects | Ideal for experiments that are sensitive to off-target events and require a high level of editing efficiency | May be beneficial for homology-directed repair (HDR) experiments, but requires two suitable cutting sites within an optimal distance of each other | |
| Molecular weight | 162,200 g/mol | |||
| Amount provided | 100 μg, 500 μg, or 5 mg | 100 μg or 500 μg | ||
| Concentration | 10 mg/mL (62 μM) in 50% glycerol | |||
| Shipping conditions | Dry ice | |||
| Storage conditions | –20°C at stock concentration | |||
| Dilution | Dilute in Opti-MEM® medium (Thermo Fisher Scientific) or PBS before use | |||
Learn more about the different Cas9 variants in the following sections:
Wild-type Cas9
The Alt-R S.p. Cas9 Nuclease V3 enzyme is a high purity, recombinant S. pyogenes Cas9. The enzymes include nuclear localization sequences and C-terminal 6-His tags. The S. pyogenes Cas9 enzyme must be combined with a gRNA to produce a functional, target-specific editing complex. For the best editing, combine the Alt-R S.p. Cas9 Nuclease V3 enzyme with the optimized Alt-R CRISPR gRNA in equimolar amounts. The enzyme is also available in glycerol-free solution.
High-fidelity Cas9
The Alt-R S.p. HiFi Cas9 Nuclease V3 offers improved specificity over wild-type Cas9, greatly reducing the risk of off-target cutting events. This Cas9 variant also preserves the high level of editing efficiency expected from a Cas9 nuclease, maintaining 90–100% on-target editing activity at most sites. For applications that are sensitive to off-target events, combining the Alt-R S.p. HiFi Cas9 Nuclease V3 with optimized Alt-R CRISPR-Cas9 gRNA (crRNA:tracrRNA) is highly recommended.
Cas9 nickases
Nickases are enzymes that produce single-strand breaks. Cas9 nickase variants have an alanine substitution within either of the 2 key catalytic domains, RuvC and HNH, of the WT Cas9 endonuclease. The RuvC mutant, D10A nickase, cuts on the targeted strand. The HNH mutant, H840A nickase, cuts the non-targeted strand. The trick to producing a DSB using nickases is to combine a Cas9 nickase – either D10A or H840A, together with two gRNA sequences-targeting sites that are close together but on opposite DNA strands. This creates a staggered DSB. For an experiment, you only need one type of nickase.
One benefit of using Cas9 nickases to introduce DSBs is that this can minimize unwanted off-target editing and support high on-target efficiency. Another reason to consider nickases in your experiments is that some Cas9 guide RNAs are less efficient than others. If you determine experimentally that there are no efficient Cas9 guides that target the site you want to mutate, then using a nickase with paired guides targeted at short distances from both sides of the desired mutation site may be a better approach.
For further information about Cas9 nickases and their use, please refer to the following articles or consult the corresponding chapter in IDT's CRISPR basics handbook:
When and how to use nickases HDR and Cas9 nickase design CRISPR basics handbook
Cas9-GFP and Cas9-RFP
The Alt-R S.p. Cas9-GFP V3 and S.p. Cas9-RFP V3 nucleases are high purity, recombinant S. pyogenes Cas9 enzymes that are expressed as fusion proteins with nuclear localization sequences and C-terminal 6-His tags. These enzymes have on-target functionality comparable to wild-type S.p. Cas9 and are designed for applications that require post-transfection visualization of the protein or enrichment of edited cells using fluorescence-activated cell sorting (FACS). These enzymes should be combined with Alt-R CRISPR gRNA in equimolar amounts.
Dead Cas9 (dCas9)
Alt-R S.p. dCas9 Protein V3 has mutations that result in the loss of nuclease activity. This protein can form RNP complexes with Alt-R gRNAs and bind to the target region specified by the gRNA without cutting the DNA. The primary use of dCas9 protein is to block transcription at a specific site on the genome. This is known as CRISPRi and is an alternative to RNAi for knockdown instead of knockout of genes.
Cas9 controls
As you plan your experiment, we recommend including positive and negative controls, so your downstream analysis will be as informative as possible. Some of the most important controls are gRNA sequences. A positive control should lead to cutting of a known target site. A negative control should not specifically cut genome sequences. Other negative controls can include samples with only vehicle, Cas protein, gRNA, or cells, as well as any other negative controls that are appropriate in your particular experiment.
When you are using Cas9 with human, rat, or mouse cells, you can use one of the Alt-R CRISPR-Cas9 Control kits. These kits contain species-appropriate positive control crRNA, negative control crRNA, tracrRNA, and PCR primers for T7EI analysis of editing from control samples. You also have the flexibility to buy kit components separately if you already have some of the items in the kit. If using sgRNA, please contact IDT for the equivalent control sequences.
Additional resources
For more detailed information about gene editing using CRISPR-Cas9, download IDT's CRISPR basics handbook. Further insights into the products can also be found on IDT's website.
Download handbook Go to IDT website Further reading and links
New in the field of CRISPR-Cas genome editing?
Get a discount on your first order!
Friendly CRISPR clash webinar
First Back2Back Academic and Industrial CRISPR Webinar in Switzerland.
- Dissecting the molecular mechanism of Cas9 specificity
Speaker: Dr. Martin Pačesa, Group of Prof. Bruno Correia, EPFL - Laboratory of Protein Design and Immunoengineering
- Improved methods for CRISPR HDR research using Alt-R™ modified donors and Alt-R HDR Enhancer V2
Speaker: Jennifer Stott, Commercial and Technical Specialist CRISPR & Functional Genomics - IDT
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend for these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.