What is CRISPR screening?
CRISPR screening is an experimental method that uses CRISPR genome editing to identify a small number of genes (out of the whole genome) involved in a certain physiological effect within cells. Applications of CRISPR screening include identifying genes associated with a disease, genes causing drug resistance in pathogenic organisms or in cancer cells, genes responsible for susceptibility to environmental toxins, and many other uses in basic research on cells.
Most CRISPR screening is done in cell culture, although some methods have been devised for use in animal models. In most CRISPR discovery experiments, scientists knock out every gene that could possibly be important for a certain phenotype, knocking out only one gene per cell. In the resulting population of cells, some cells die, while others survive and may even be able to grow better, becoming the predominant cell type. Then, scientists perform next-generation sequencing (NGS) on the final population of cells to determine the gene responsible for the observed phenotype.
There are viral (pooled) and non-viral (arrayed) approaches to CRISPR screening. Both have their advantages and disadvantages, and the choice of method depends on the specific research question and experimental setup. Read more about this topic in the following sections and discover our products that support you in your research. To receive a more detailed overview of CRISPR screening, please refer to the article from our supplier Integrated DNA Technologies (IDT).
Article: What is CRISPR screening? Learn more about CRISPR genome editing
Pooled approach to CRISPR screening
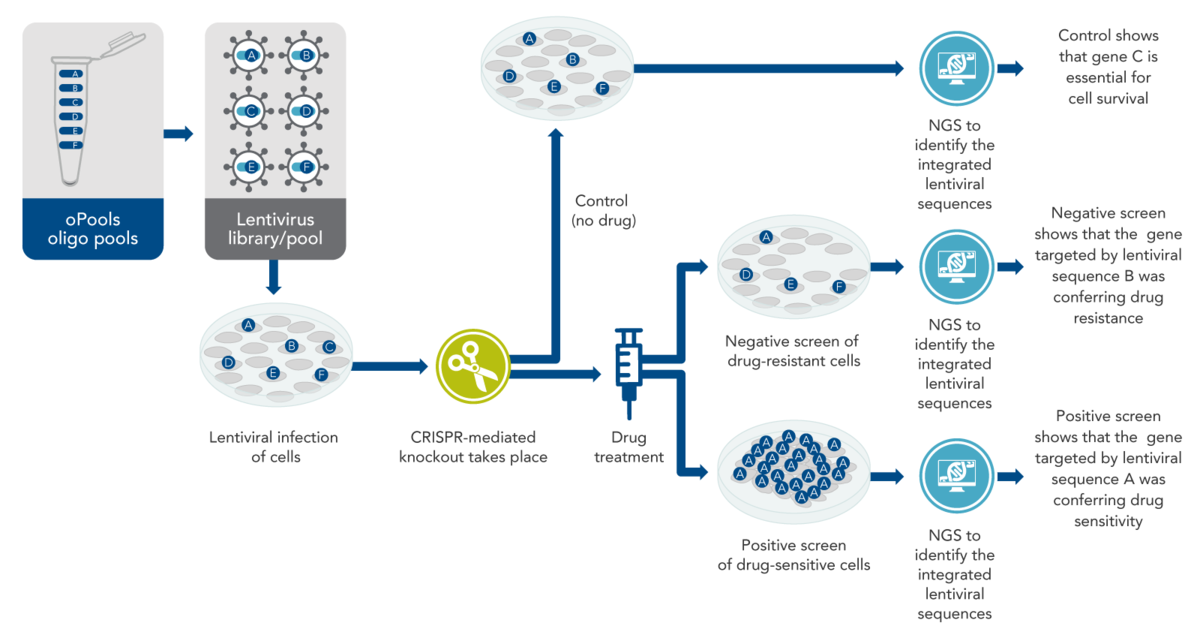
For the commonly used pool approach, each oligo in the pool contains DNA to encode at least the targeting region of the CRISPR guide, or frequently the entire single-guide RNA (sgRNA) including the target sequence. Each oligo also must have sites at each end to allow cloning into lentiviral gene-containing plasmids appropriately designed for biosafety. From the plasmids, lentiviruses are produced as a pool containing thousands of CRISPR-targeting sequences, with one targeting sequence per virus particle (virion).
Lentiviral libraries are used to infect cells (goal: no more than one virion per cell). After infection, lentiviral RNA is first reverse-transcribed to DNA, which in turn is integrated into the genome of infected cells. After integration, the lentiviral sequences—including the cloned-in CRISPR sequences—are transcribed to RNA, leading to the production of the guide RNAs. Methods to deliver Cas enzymes into cells for lentiviral CRISPR screening include using a cell line that stably expresses a Cas enzyme or producing a pool of lentiviruses that contain both the gene for the desired Cas enzyme and the DNA that codes for the guide RNA.
After treatment of the cells with the viral library of targeting sequences and the desired Cas enzyme, the cells are incubated for a few days. This allows the cells time to develop phenotypic CRISPR-mediated changes—in many experiments, this means the cells either grow or die off. Then, drug or other treatment is performed if desired for the specific experiment. After this, total genomic DNA samples or total RNA samples from the two mixed populations (i.e., the control cells and the drug-treated cells) are collected for sequencing. The DNA (or RNA) is subjected to sequencing by NGS. The two resulting lists of sequences (control vs. drug-treated) can then be compared.
oPools™ Oligo Pools
Use oPools Oligo Pools for accurate, reliable, and affordable CRISPR libraries. These pools of custom single-stranded DNA sequences offer high fidelity, uniformity, low error rates, and low dropout rates. This means you can avoid amplification bias, varying concentrations, or high error rates.
Specifications | |
|---|---|
| Length | 40–350 bases/oligo |
| Scale (number of oligos) | Up to 20'000 |
| Mixed bases | N, R, Y, M, K, S, W, H, B, V, D |
| Modifications | 5′ phosphorylation |
| Shipping container | 2 mL tube |
Endura™ Competent Cells for CRISPR library construction
Endura Competent Cells are a commonly used strain for cloning sequences that suffer unwanted recombination events in other strains. Clone repetitive sequences and lentiviral CRISPR libraries with high efficiency.
- Stabilize direct repeats and generate lentiviral guide RNA libraries
- Choose electrocompetent or chemically competent cells
- Highest efficiency commercially available cells for lentiviral cloning: over 1 × 107 cfu/μg (chem) or 1 × 1010 cfu/μg (electro)
Next-generation sequencing services
LubioScience offers comprehensive services for next-generation sequencing in collaboration with accredited state-of-the-art laboratories.
CRISPR screening using arrayed gRNAs
Traditionally, almost all CRISPR screening was done with a pool of lentiviruses carrying a large number of sequences to be targeted. However, scientists may already have a short list of genes of interest and they want to verify a narrower set of targets that were identified in an initial lentiviral primary screen, or they may require a more sophisticated genotype-to-phenotype screening workflow. In either instance, a different approach can be taken:
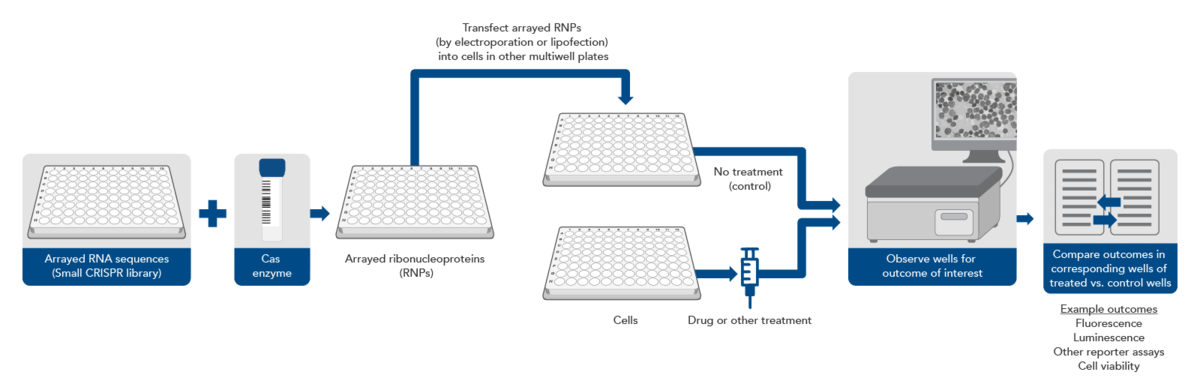
A synthetic gRNA CRISPR library consists of synthetic RNA guides kept in separate wells in an array format on a multi‑well plate. The most common array formats are a single guide per well or multiple guides per gene, pooled together by gene, so that each well will knock out a single gene. In this experimental design, gRNAs can be complexed with an appropriate Cas enzyme in each well. These ribonucleoprotein complexes can be transfected, by electroporation or lipofection, individually into cells in different wells of a multi-well plate. Thus, each well of cells is transfected with one or several predetermined CRISPR targeting sequences. Then, depending on the experimental setup, the wells can be monitored for a physiological effect.
This approach eliminates the need for NGS but requires automation equipment to avoid being extremely labor intensive and can be much more expensive if a large library is used. However, by removing the need for lentivirus, as few as 2–3 target sites per gene need to be selected for high editing efficiency, as there is no chance that lentiviral integration will disrupt an important genomic sequence. Avoiding lentivirus also removes biosafety requirements and ensures that the synthetic gRNAs will not continue to exist in the cells after the knockout has occurred, decreasing the likelihood of off-target editing effects. Additionally, this approach is more appropriate for sophisticated screens where the parameters are around change in phenotype caused by the gene knockout, which can be viewed through microscopy, rather than just live or dead cells.
More information about CRISPR functional genomics screens using arrayed guide RNA libraries can be found in the following article.
CRISPR gRNA Libraries
Alt-R™ CRISPR gRNA Libraries are custom arrayed synthetic guide RNA libraries. They are a great way to accomplish CRISPR knockout screens and accelerate your research by avoiding the cloning and sequencing associated with lentiviral screens, while also integrating into existing automation workflows. IDT has experience designing and manufacturing arrayed panels of gRNAs in a range of sizes, from a few guides to thousands or more.
Your benefits:
- Highly customizable CRISPR libraries: Different gRNA formats (2-part or sgRNA), plate type, custom formulations, and lab automation platforms, to suit a variety of project needs
- Reliable, consistent solution offered by a global leader in RNA synthesis and innovation
- Adaptable for alternative CRISPR systems such as Cas12a, Cas13, and prime editing
- Enhanced nuclease resistance for maximal editing in Cas nuclease-expressing cells or via ribonucleoprotein delivery
Additional resources
For more detailed information about gene editing using CRISPR-Cas, download IDT's CRISPR basics handbook. Further insights into IDT's products for CRISPR genome engineering can also be found on their website.
Download handbook Go to IDT website Further reading and links
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend for these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.
