What is the rhAmpSeq™ CRISPR Analysis System?

The rhAmpSeq™ CRISPR Analysis System is an advanced data analysis pipeline from Integrated DNA Technologies (IDT) for quantifying your genome editing results. Expect quick and accurate quantification of CRISPR-Cas edits. IDT's proprietary RNase H2-dependent PCR technology generates amplicon libraries for targeted sequencing on Illumina® NGS platforms. The system includes an advanced, yet accessible, cloud-based data analysis pipeline for quantification of on- and off-target edits.
Your advantages with this CRISPR Analysis System
- Best-in-class insertion/deletion (indel) quantification of genome editing
- Batch analysis of your results (multiplex up to 500 targets)
- High sequencing coverage uniformity and percentage of mapped reads
- Intuitive data analysis that requires no bioinformatics expertise
- Sample-to-analysis in under a week
Harnessing the power of rhAmp™ PCR for characterizing on- and off-target editing
The rhAmpSeq CRISPR Analysis System depends on IDT's proprietary rhAmp™ PCR technology. Using RNA-base-containing blocked primers (rhAmp primers), this technology harnesses the intrinsic specificity of the RNase H2 enzyme to cleave DNA:RNA duplexes. Due to its inherent mitigation of primer dimers, rhAmp PCR exhibits high specificity, even in multiplex reactions. The rhAmpSeq system combines this innate advantage with a convenient workflow that requires only two PCR steps (Figure 1) to generate amplicon libraries for Illumina® sequencing platforms. Figure 1 shows both amplification steps in the rhAmpSeq workflow, highlighting the role of rhAmp PCR technology during the first step (Targeted rhAmp PCR 1). Coupled with IDT's industry-leading oligo manufacturing process, the rhAmpSeq system offers fast and cost-effective analysis of your CRISPR results. Advantages of rhAmp PCR include an increased target affinity compared to traditional PCR, less off-target amplification, and a reduction of primer-dimers.
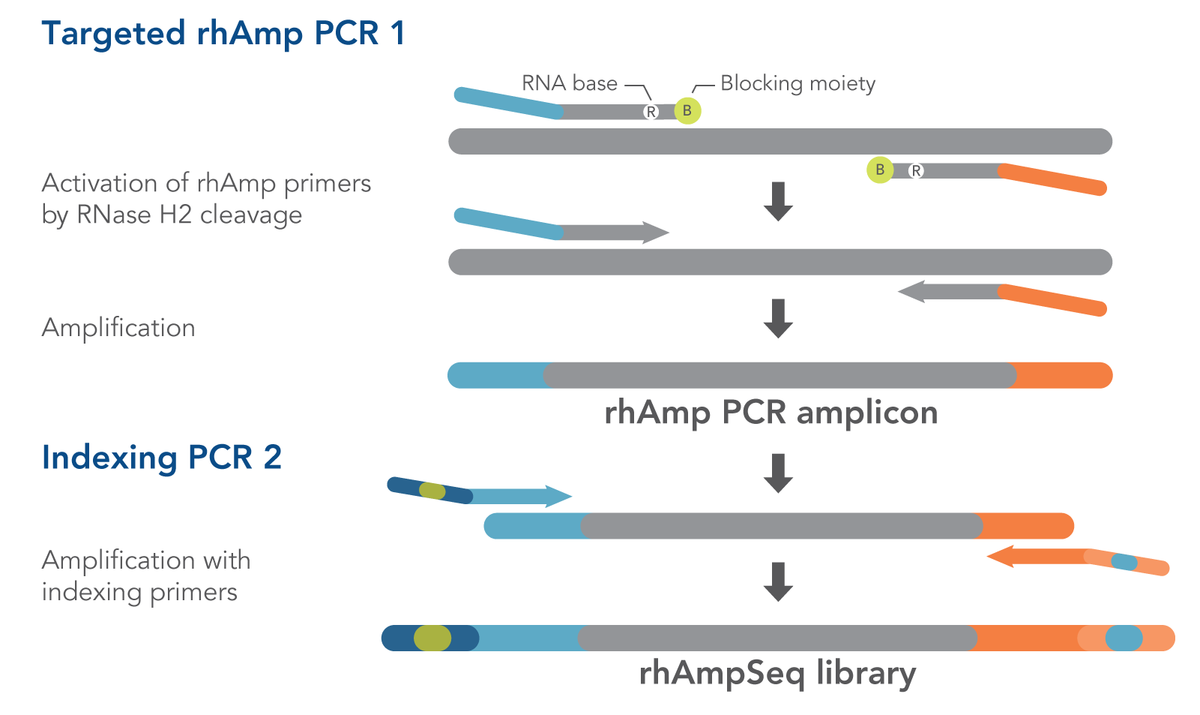
Workflow for analyzing your CRISPR-Cas editing results
Before you begin: Identify and nominate both on- and off-target sequences. The rhAmpSeq Analysis System workflow begins after the identification of potential targets. Off-target sites can be nominated using empirical methods such as GUIDE-Seq with or without in silico prediction tools.
Additional ressources
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend for these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations.
