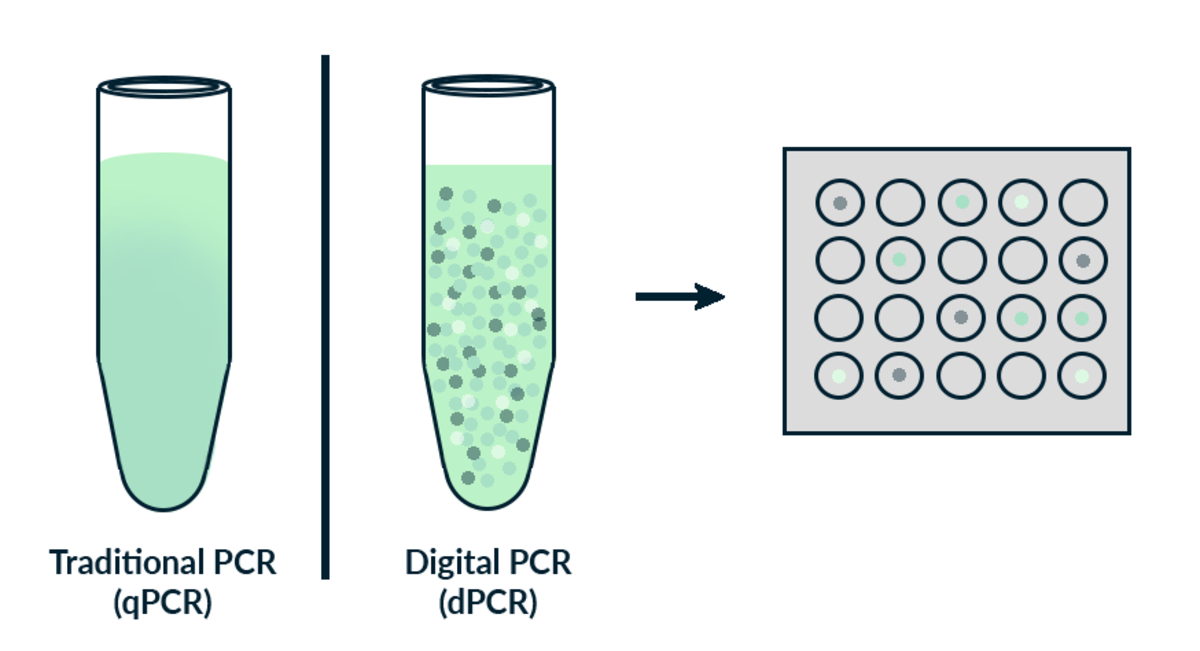
Digital PCR (dPCR) is the third generation of PCR after end-point PCR and real-time quantitative PCR (RT-qPCR). Digital PCR offers high sensitivity, accuracy and the ability to absolutely quantify the amount of target DNA in a sample. This high level of sensitivity permits detection of rare mutations, copy number variations (CNV), low abundance transcripts, rare microRNAs and very low viral loads.
Fundamental principles
dPCR differs from standard endpoint-PCR in that the sample is first partitioned randomly into 20’0000 sub reactions, rather than running as a single reaction on a bulk sample. The PCR reaction is performed on each partition either in individual droplets or on a microfluidic chip. Ideally, each partition will contain either one or zero target sequence. In order to compensate for the few partitions that will contain more than one target sequence, the data is treated by means of Poisson statistics. If the partition contains an amplifiable sequence it is counted as positive (otherwise it is counted as negative).
Assuming a single target per partition allows absolute quantification of how many targets were in the original sample by counting the number of positives and extrapolating based on the number of partitions and dilution factor. As the actual number of targets are counted, a calibration curve is not required, although is it recommended to run controls of known samples and quantities to aid in the validation of data, particularly for publication.
Workflow
1. Sample preparation
As with any genomic analysis, good sample preparation is essential. dPCR can be used to amplify any purified DNA sample (gDNA or cDNA) but as with qPCR is unable to amplify RNA directly. Any mRNA targets of interest must therefore be isolated and converted to cDNA prior to dPCR analysis.
Purified gDNA or cDNA is then mixed with forward and reverse primers, along with a hydrolysis probe, each of which is designed to have sequence complementary to the target gene/DNA of interest. As with classical PCR, primers anneal to the 3’ and 5’ ends of the target sequence (typically 60–150 bps in length). In addition, and different to classical PCR, the 5′ nuclease (hydrolysis) probe then anneals between the forward and reverse primers and includes a 5′ fluorophore and 3′ quencher. The hydrolysis probes increase sensitivity of the reaction. As with all PCR and qPCR in particular, primer and probe design is key to obtaining good data.
IDT offer a variety of different assays to perform dPCR, including PrimeTime™ qPCR Probes, PrimeTime qPCR Probe Assays, or Affinity Plus™ qPCR Probes with locked nucleic acids (Tables 1 & 2).
Digital PCR can also be done using SYBR® Green (Life Technologies) or EvaGreen® (Biotium) intercalating double-stranded DNA dyes rather than 5′ nuclease probes.
2. Dilution and partition
Following sample preparation and addition of primers and master mix, dPCR assays partition the sample mix into individual nanolitre reactions so there is either 1 or 0 target DNA molecules in each partition. Some dPCR platforms divide the mix into individual micro moulded plastic wells, whereas other dPCR platforms create nanolitre sized reaction droplets as an oil/water emulsion. dPCR then analyses around 20,000 individual qPCR reactions.
3. PCR amplification and end-point analysis
Following partitioning, reactions are amplified using regular PCR cycling parameters. As DNA polymerase extends from the forward primer, its exonuclease activity degrades the probe, releasing the 5′ fluorophore from the 3′ quencher, which in turn emits detectable fluorescence. If the assay includes intercalating dyes rather than a hydrolysis probe, then the fluorescence increases as double-stranded PCR amplicons accumulate.
After the PCR cycle is complete, any partition that included one template or target DNA sequence will fluoresce. The total number of droplets or wells with fluorescence represents the total number of target molecules in the sample. Once experimentation is complete, data can be analysed via standard tools.
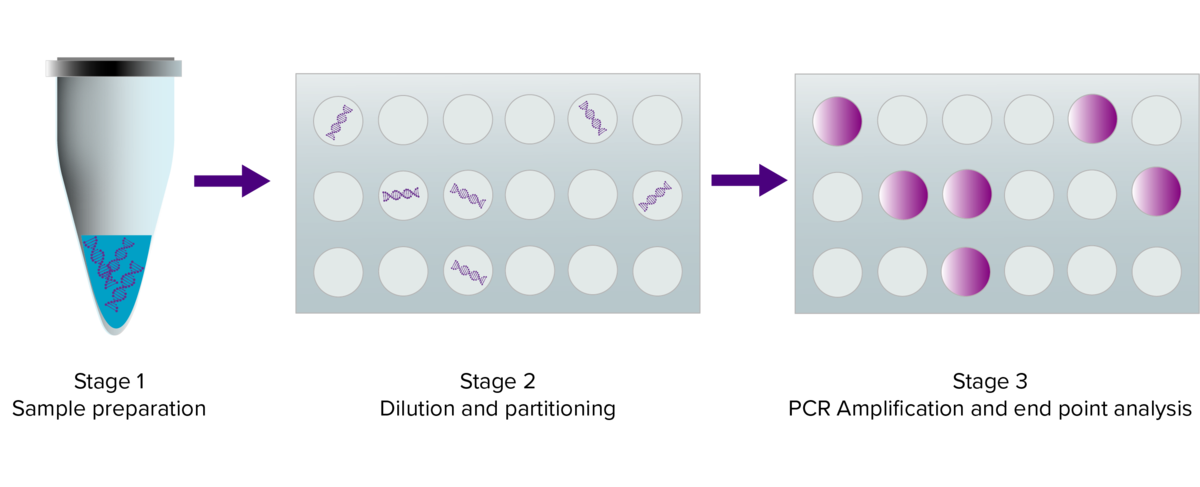
Figure 1: Digital PCR workflow
Stage 1 – gDNA, cDNA isolation and purification, addition of forward and reverse primers, hydrolysis proves or intercalating double-stranded DNA dyes.
Stage 2 – Sample partitioning into individual nanolitre reactions containing 1 or 0 target DNA molecules.
Stage 3 – PCR amplification and analysis
qPCR probes and assays for various applications
IDT offer a variety of probes and assays for genotyping and gene expression applications using microwell or droplet-based dPCR platforms:
SNP Genotyping applications
Generate reliable, high-quality gene expression analysis data with double- and single-quenched fluorescent hydrolysis probes.
PrimeTime™ qPCR Probes are 5′ nuclease probes, available with an assortment of reporter-dye combinations that are compatible with common qPCR instruments and that include several license-free combinations.
- Select from a wide range of license-free dyes, including FAM, Yakima Yellow®, SUN™, HEX, Cy® 3, Texas Red®-X, and Cy® 5 dyes
- Reduce background signals, even with longer probes, when using ZEN™ or TAO™ Double-Quenched Probes
- Successfully multiplex with reduced crosstalk using double-quenched probes
- Choose from the convenient 0.5–2.5 nmol packaging size for digital PCR (dPCR) platforms
- Achieve high efficiency of qPCR under fast or standard cycling conditions
- Minimize troubleshooting: PrimeTime qPCR Probes are verified by mass spectrometry* and HPLC-purified
* With the exception of mixed base oligos, which could potentially represent multiple sequences and therefore cannot be accurately evaluated by ESI mass spectrometry.
Table 1: Mini Affinity Plus™ qPCR Probes
| 5’ reporter dye(s) | Emission (nm) | Quencher(s) | Delivery amount |
|---|---|---|---|
| FAM | 520 | Iowa Black FQ | 0.5 nmol |
| SUN™ | 554 | Iowa Black FQ | 0.5 nmol |
| HEX | 555 | Iowa Black FQ | 0.5 nmol |
| Cy®5 | 668 | Iowa Black RQ | 0.5 nmol |
Gene Expression applications
Primer and probe premixed assays for analyzing gene expression in any species using fluorescently labeled 5′ nuclease probes
PrimeTime™ qPCR Probe Assays consist of a primer pair and fluorescently labeled 5′ nuclease probe. Obtain predesigned assays for human, mouse, or rat for easy selection based on multiple criteria such as exon location and number of transcripts detected. Create custom assays for any sequence from any species using the PrimerQuest™ Tool.
- Achieve >90% efficiency over 4 or more orders of magnitude with our predesigned assays, guaranteed
- Select from a wide range of license-free dyes, including FAM, TET, SUN™, HEX and Cy® 5 (Cytiva) dyes
- Multiplex with confidence using double-quenched ZEN™ or TAO™ probe assays that have the lowest background in the industry
- Select a primer:probe ratio from 2:1 to 4:1 with no additional charge
- Size and scale options available for digital PCR (dPCR) platforms
- Receive primers and probe sequences with all orders
Table 2: PrimeTime™ qPCR probes and assays
PrimeTime™ qPCR Probes – small scale synthesis
| 5’ reporter Dye | Emission (nm) | Quencher(s) | Delivery amount Mini (0.5 nmol) | Delivery amount Eco (2.5 nmol) | Minimum guarantee Other scales (nmol) |
|---|---|---|---|---|---|
| FAM | 520 | ZEN™/Iowa Black FQ | ✓ | ✓ | 15, 25, 50 |
| SUN™ | 554 | ZEN™/Iowa Black FQ | ✓ | ✓ | 15, 25, 50 |
| HEX | 555 | ZEN™/Iowa Black FQ | ✓ | ✓ | 10, 25, 50 |
| Cy®5 | 668 | TAO™/Iowa Black RQ | ✓ | ✓ | 2, 8, 20 |
PrimeTime™ Probe Assays in tubes
| 5’ reporter Dye | Emission (nm) | Quencher(s) | Unit size | Reactions | Other reaction sizes |
|---|---|---|---|---|---|
| FAM | 520 | ZEN™/Iowa Black FQ | Mini | 100 | 500, 2500 |
| SUN™ | 554 | ZEN™/Iowa Black FQ | Mini | 100 | 500, 2500 |
| HEX | 555 | ZEN™/Iowa Black FQ | Mini | 100 | 500, 2500 |
| Cy®5 | 668 | TAO™/Iowa Black RQ | Mini | 100 | 500, 2500 |
PrimeTime™ Probe Assays in 96-well plates
| 5’ reporter Dye | Emission (nm) | Quencher(s) | Unit size | Reactions | Other reaction sizes |
|---|---|---|---|---|---|
| FAM | 520 | ZEN™/Iowa Black FQ | Mini | 100 | 500, 2500 |
Multiplex digital PCR
The individual partitions are also amenable to multiplexing where multiple target DNAs or cDNAs are analyzed simultaneously. Just like a multiplex qPCR reaction, 2–5 sets of primers and probes with distinct fluorophores are mixed with the sample. Since the template or target DNAs are partitioned into individual reactions, one set of primer and probes will amplify the individual target, and therefore, the primers and probes will not compete for reagents during amplification as in a typical multiplex qPCR assay. During analysis, the number of partitions for each of the fluorophores represents the number of individual targets for that specific primer and probe set. Counting the partitions for each primer and probe set in the multiplex assay can establish an absolute ratio for the different targets. Multiplexing dPCR reactions are sensitive to detect even rare alleles for a gene and determine copy number variations.
Who is IDT?
Integrated DNA Technologies (IDT) is your advocate for the genomics age. They produce tools for NGS, CRISPR, qPCR and PCR. Their products include DNA/RNA oligos, genes and gene fragments. For more than 30 years, IDT's innovative tools and solutions for genomics applications have been driving advances that inspire scientists to dream big and achieve their next breakthroughs.