The Problem
DNA-targeting CRISPR-Cas enzymes are limited to targeting a subset of sequences within the genome because they require protospacer-adjacent motif (PAM) recognition. This limits the utility and affects the editing efficiency and flexibility of CRISPR-Cas gene editing systems. PAM requirement prevents accurate positioning of CRISPR target sites and is a barrier for high-resolution target positioning.
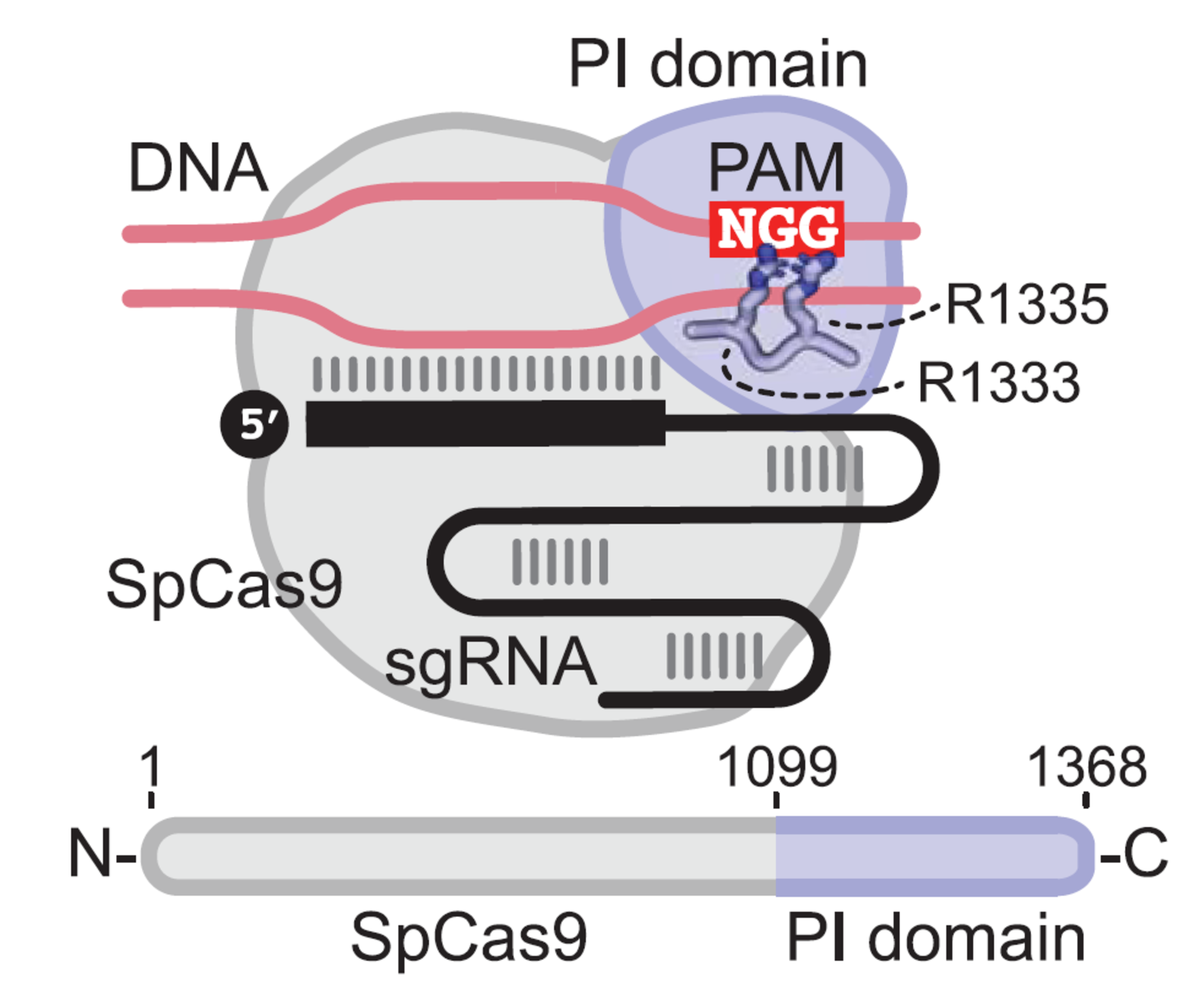
What is a PAM?
Protospacer-adjacent motifs (PAM) are short sequence motifs adjacent to target sites in foreign DNA that help CRISPR systems distinguish self from non-self. Cas9 from Streptococcus pyogenes (SpCas9) recognizes target sites with NGG PAMs, where N is A, C, G, or T, using its PAM-interacting domain – R1333 is particularly important for recognition.
Walton, et al., generated and optimized variants of Cas9 enzymes to eliminate the NGG PAM requirement, expanding the set of recognizable PAMs and targetable DNA sequences.
Relaxing SpCas9 PAM preference
The researchers created a variant of SpCas9 that recognizes an NGN PAM (where N is any of the four DNA bases) by using SpCas9-VRQR as a molecular scaffold, with VRQR corresponding to the following amino acid substitutions: D1135V/G1218R/R1335Q/T1337R. Using an in-house-developed high-throughput determination assay (HT-PAMDA), the scientists profiled PAM preferences of SpCas9 variants and determined the contributions of substitutions at key PAM-identifying positions: D1135, S1136, G1218, E1219, R1335, and T1337.
The SpCas9-LWKQQR variant, dubbed SpG, evenly targeted NGN PAMs in the HT-PAMDA and HEK293T cells, successfully relaxing PAM requirements. And SpG targeted NGN PAMs more evenly and robustly compared to nucleases SpCas9-NG and xCas9 in human cells and HT-PAMDA.
SpG base editing activity
Base editing technologies mediate single nucleotide substitutions. Walton, et al., generated BE4max cytosine base editor (CBE) constructs of SpG, SpCas9-NG, wildtype Sp-Cas9, and xCas9, all harboring similar PAM profiles as their corresponding nuclease constructs.
They found SpG- and SpCas9-NG-CBE constructs were more efficient than WT- and xCas9-CBE, with a mean C-to-T editing efficiency of >23% across NGN PAMs (mean C-to-T editing efficiency of WT- and xCas9-CBE >15% at NGG PAMs). Similarly, SpG and SpCas9-NG adenine base editor (ABE) constructs efficiently generated A-to-T substitutions as NGN PAMs, with SpG-ABE being the most robust.
Generating a near-PAMless SpCas9
To further relax PAM requirements and make more of the genome accessible to genome editing, SpG served as a scaffold. The goal was to alter recognition of the second position of the NGN PAM. First, the researchers generated SpG-R1333Q in the hopes it would recognize NAN and NGN PAMs, but it unfortunately abolished activity in cells. They rescued nuclease activity in SpG-R1333Q by introducing two additional mutations: L1111R and A1322R, which without R1333Q negatively impacts SpG activity.
Using SpG-L111R/A1322R as a scaffold, the researchers tested all 20 amino acids at R1333 to evaluate differences in PAM recognition and nuclease activity. Alanine, cysteine, and proline substitutions at 1333 demonstrated efficient NRN PAM targeting (where R is A or G) compared to R1333Q.
The team continued tinkering with various residues in the nuclease to generate more nonspecific DNA contacts and maximize PAM flexibility. They found SpG(L111R/A1322R) harboring R1333P, A61R, and N1317 produced the most robust average editing efficiency in cells at NRN PAMs and can even target NYN PAMs, where Y is cytosine or thymine. This near-PAMless variant was named SpRY.
SpRY base editing activity
In HEK293T cells, SpG remained the most efficient NGN PAM-targeting variant and SpRY was more effective at NRN PAMs compared to wildtype, except for sites with canonical NGG PAMs. Additionally, SpRY targeted NYN PAMs in human cells at ~50% mean editing activity compared to NRN PAMs, and the reason for preference needs to be further explored.
The researchers tested SpRY CBE and ABE activity and compared it to WT. They found that SpRY is a more efficient base editor compared to WT at all sites, except those harboring an NGG PAM. Furthermore, editing activity was robust at high-activity NRN and NYN sites revealed during nuclease testing.
Improving fidelity and specificity
Off-target effects are always a concern when using genetic engineering technology. While relaxing PAM requirements made more of the genome targetable, it increased off-target effects. The researchers successfully eliminated nearly all off-target effects by generating high-fidelity constructs of SpG and SpRY.
Proof of concept
The researchers demonstrated the utility of SpG and SpRY in a proof-of-concept experiment where they generated genetic variants thought to be protective against coronary heart disease, type 2 diabetes, osteoporosis, chronic pain, and various pathologies in human cells. Wildtype SpCas9 successfully edited at NGG PAM sites, but it produced potentially deleterious bystander editing of nearby cytosines, whereas this was avoided with SpG- and SpRY-CBEs targeting nearby NGC and NAT PAMs.
At NRN PAMs lacking nearby canonical NGG PAMs, WT-CBE was inefficient, demonstrating the need for expanded targeting. By having more targetable PAM options, SpG- and SpRy-CBEs generated the intended base edit across all targeted sites, where wildtype fell short, without introducing detrimental bystander editing.
The impact
Walton et al., produced Cas9 variants that enables researchers to target areas of the genome that were previously inaccessible, while enhancing editing resolution and preserving specificity. This research specifically looked at Cas9, but the strategy employed can be applied to various Cas enzymes to generate a massive toolkit of gene editors.
References
Walton et al., "Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants.” Science 368, 290–296 (2020). DOI: 10.1126/science.aba8853