Deoxyribonucleic acid (or DNA) is one of the most important macromolecules in biology. It plays a central role in many critical cell processes such as protein synthesis and is the blueprint for much of cellular structure.
Because of DNA's importance, many visualization and quantification tools have been developed. Reagents and probes have been created to detect DNA in all kinds of applications- microscopy, gel electrophoresis and flow cytometry to name a few.
With over 50 different dyes on the market, it can be overwhelming to determine the optimal dye for any given experiment. To help you choose, here's what we consider to be the best DNA stains and probes.
Ethidium bromide
Pros & Cons
+ Cheap
+ Well-established, numerous protocols
- Potential hazard, mutagenic
- Costly to dispose of
Applications
Gel electrophoresis, fluorescence microscopy, quantitation
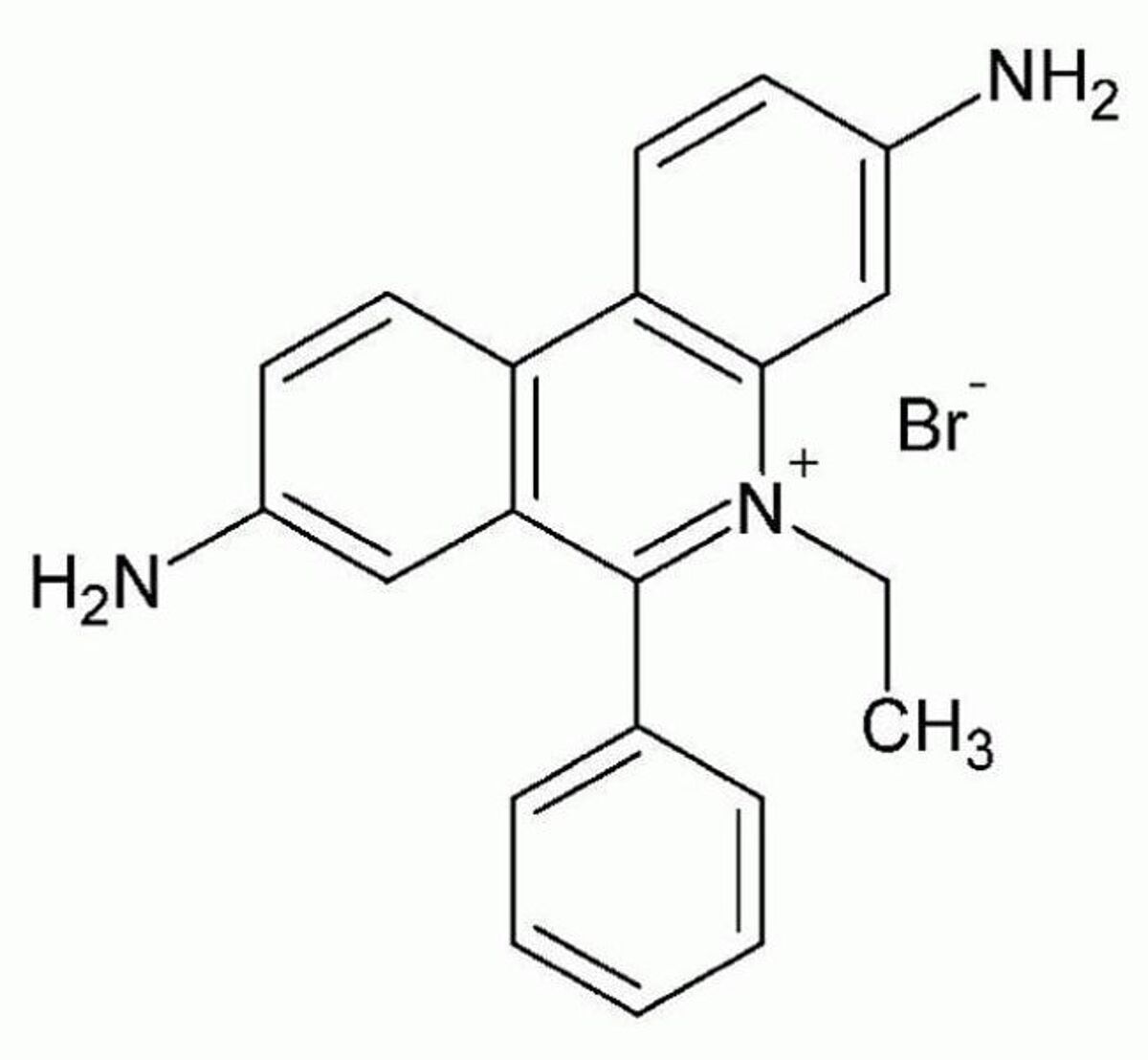
Ethidium bromide (EtBr) is one of the most popular DNA probes. This is because it was one of the first to be commercially available: as early as the 1950s, it was used to treat diseases in livestock. In the 1970s, scientists began using it as a DNA probe. In addition to its early adoption, ethidium bromide is comparatively inexpensive. Even in large quantities, it remains very affordable, running around a few Swiss Francs per gram.
Upon binding to DNA, EtBr experiences a roughly 20-fold increase in brightness. It releases an orange fluorescence (605 nm) when excited by UV light (~300 nm). Ethidium bromide can also detect RNA depending on how much RNA folding occurs. Ethidium bromide is not a good probe for detecting DNA in live cells, however. This is because it is impermeable to intact cell membranes.
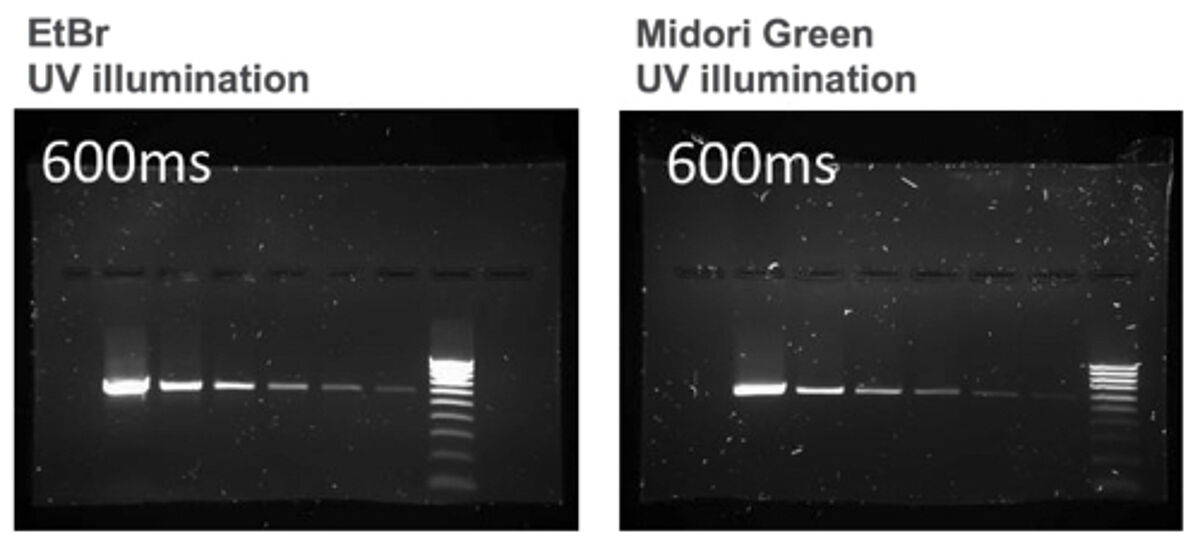
Over the years, health concerns have arisen over ethidium bromide usage. In particular, concerns have been raised about ethidium bromide's role as a mutagen. Ethidium bromide can interfere with DNA replication and transcription in humans when exposed in large quantities. This is due to its properties as an intercalating agent. At low concentrations, however, it is not considered a hazardous waste. This has spurred intense debate as to whether ethidium bromide is optimal for conventional laboratory use and has led to the adoption of alternative, safer dyes such as Midori Green (see below).
There are a lot of safe nucleic acid stains on the market and Midori Green is certainly one of the best out there. It features a full safety profile, including mutagenic testing and glove penetration tests, and can be disposed down the sink without any additional measures. Pricewise, its high dilution factor (1:50'000 for agarose gel staining) makes it a very economical choice for nucleic acid staining. There are multiple formulations of Midori Green, which have been optimized for direct addition to DNA or RNA, as well as for in-gel staining and post-staining. An additional advantage is that Midori Green works both with your old UV table and modern, safer Blue/Green light tables.
Propidium iodide
Pros & Cons
+ Well established for flow cytometry
- Potential hazard, mutagenic
Applications
Flow cytometry, multiplexing, microscopy
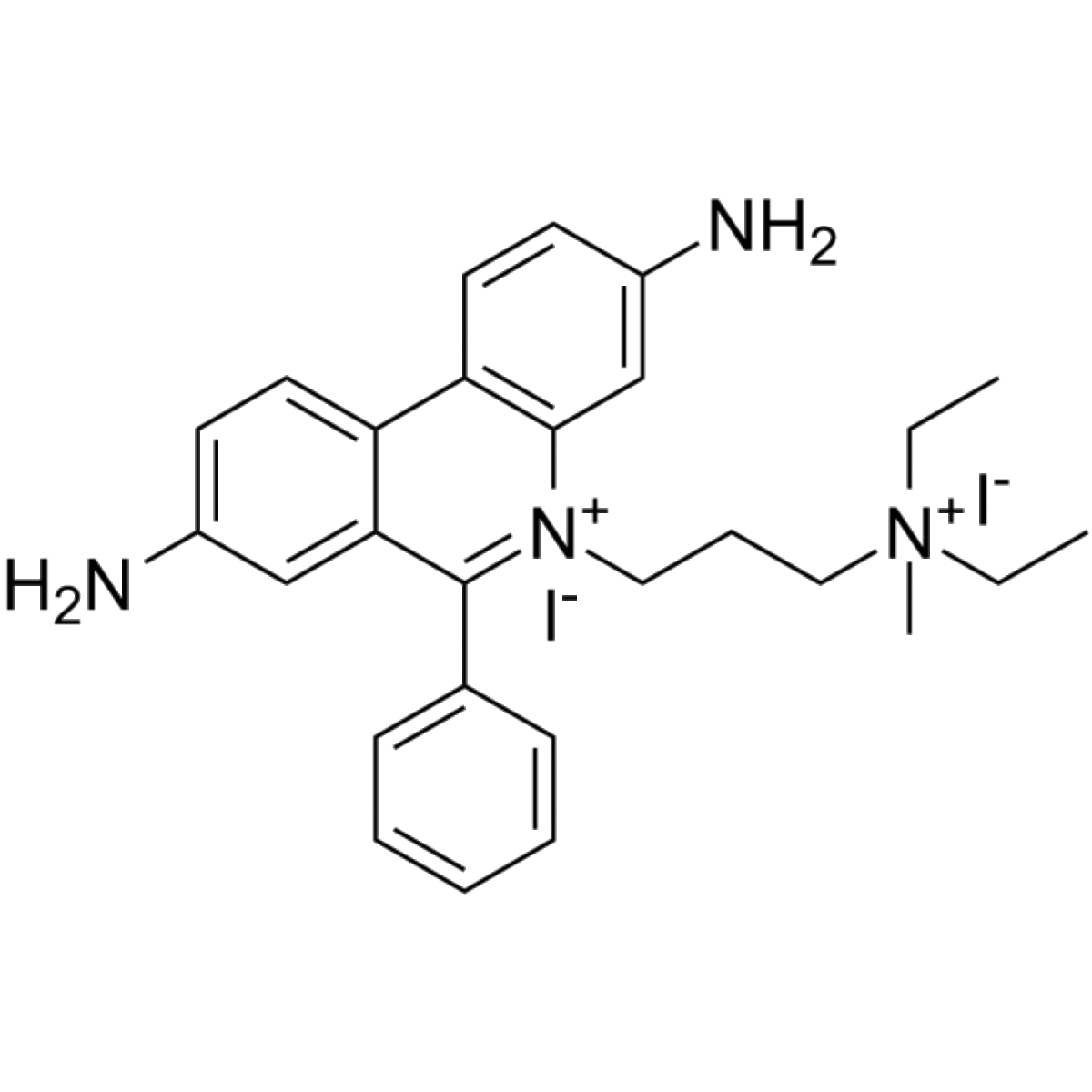
Propidium iodide is a DNA probe and an intercalating agent. It is in the same chemical family as ethidium bromide. Like ethidium bromide, propidium iodide has a nitrogen-containing ring structure that forms its core but it differs in that it has an additional quaternary amine which binds ionically to an iodide ion. Upon binding to DNA, propidium iodide will experience a 20-30 fold increase in fluorescence and it is also able to bind to RNA if folding occurs.
Propidium iodide is membrane impermeable and only enters cells with compromised membranes, which makes it an excellent probe for identifying dead cells. It can also be used to quantitatively assess DNA content in a biological sample.
Propidium iodide shows no sequence preference and binds roughly once per 4-5 DNA base pairs. It can be excited by a xenon- or mercury- lamp as well as a 488 nm argon-ion laser and because of its emission at 617 nm, it can easily be used in multiplex assays with green fluorescent probes such as fluorescein. It can also be used as a counterstain for multi-color analysis. Propidium iodide is most commonly used in flow cytometry applications.
dUTP-conjugated Probes
Pros & Cons
+ Versatile
+ Excellent probe for FISH
Applications
Direct labeling, hybridization, PCR, FISH, DIG-dUTP
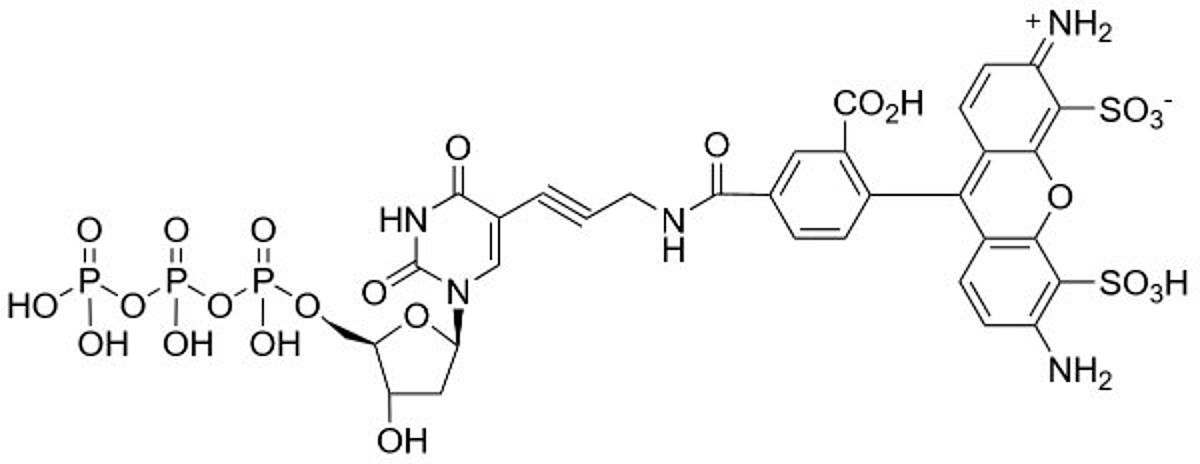
dUTP-conjugated probes form an interesting class of DNA detectors mainly resulting from the experimental flexibility they provide. The basic idea is to attach a probe to dUTP with what's called a linker. Then, the dUTP is incorporated into DNA through molecular techniques. This also incorporates the probe into the DNA macromolecule. In this way, the DNA becomes labeled with the probe.
Because dUTP can be conjugated with many different probes, dUTP-conjugates have a wide range of potential applications. For example, dUTP probes can be used to monitor PCR reactions. They can also be used as the probe in FISH procedures. In general, dUTP probes excel in situations where DNA hybridization takes place.
Some of the most common dUTP conjugates include labeling with compounds like digoxigenin (DIG), biotin and fluorescein. Labeling with these compounds all provide non-radioactive probes for DNA detection. These probes in particular have also found widespread use in immunoassays like ELISA.
| Cat-No. | Item | Size | Price (CHF) |
|---|---|---|---|
| 17023 | Tetramethylrhodamine-dUTP *1 mM in TE Buffer (pH 7.5)* | 25 nmol | 163.00 |
| 17011 | mFluor Violet 450-dUTP *1 mM in TE Buffer (pH 7.5)* | 25 nmol | 433.00 |
| 17043 | iFluor647-PEG12-dUTP *1 mM in TE Buffer (pH 7.5)* | 25 nmol | 151.00 |
| 17042 | iFluor555-PEG12-dUTP *1 mM in TE Buffer (pH 7.5)* | 25 nmol | 151.00 |
| 17006 | TF1-dUTP *1 mM in TE Buffer (pH 7.5)* | 25 nmol | 433.00 |
| 17007 | TF2-dUTP *1 mM in TE Buffer (pH 7.5)* | 25 nmol | 433.00 |
| 17008 | TF3-dUTP *1 mM in TE Buffer (pH 7.5)* | 25 nmol | 433.00 |
| 17009 | TF4-dUTP *1 mM in TE Buffer (pH 7.5)* | 25 nmol | 433.00 |
| 17010 | TF5-dUTP *1 mM in TE Buffer (pH 7.5)* | 25 nmol | 433.00 |
| 17040 | XFD488-dUTP *1 mM in TE Buffer (pH 7.5)* | 25 nmol | 277.00 |
DAPI
Pros & Cons
+ Gold standard for staining nulceic DNA in microscopy
+ Well-established, many protocols
- Cytotoxic
Applications
Fluorescence microscopy
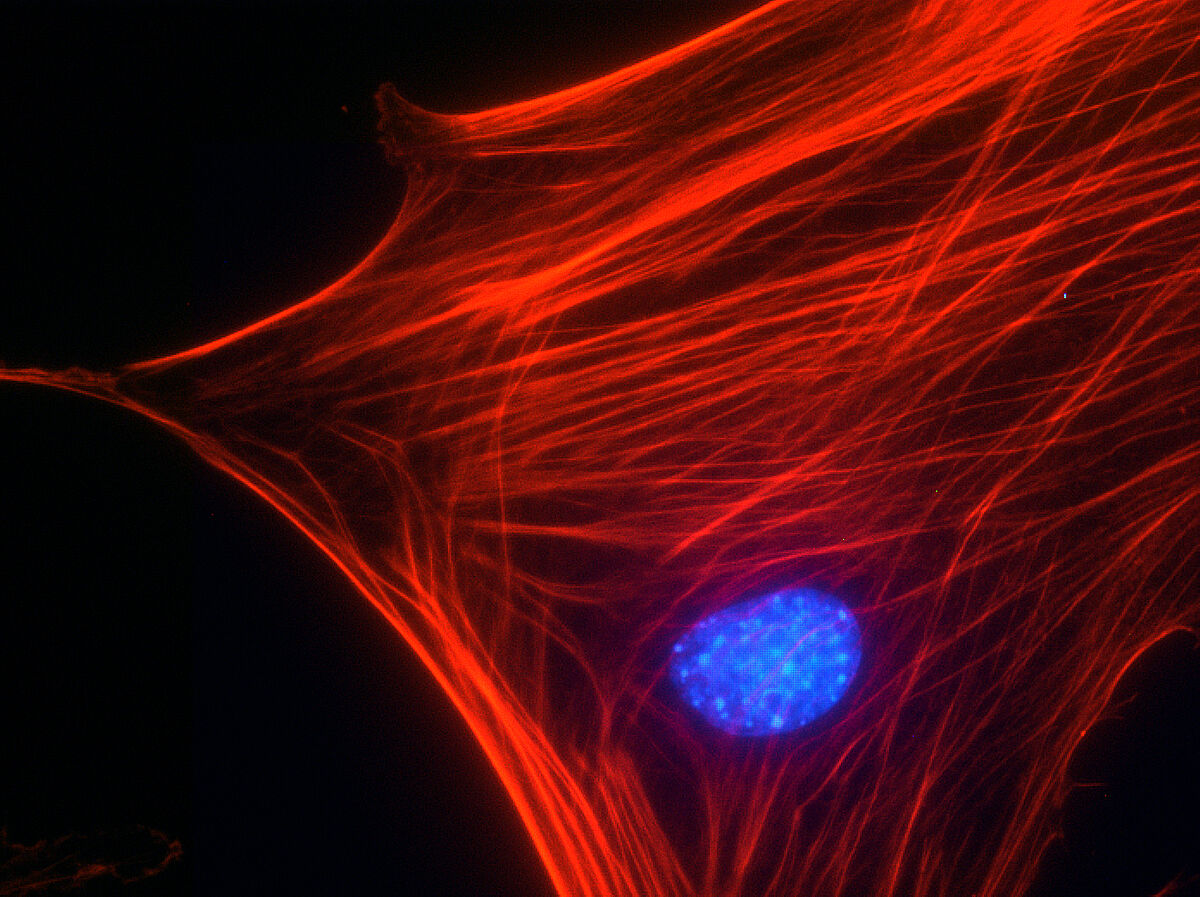
DAPI, like many of the dyes on this list, was first synthesized as a medical agent. It was initially used in an attempt to treat trypanosomiasis, a disease caused by parasites. In the late 1970s, DAPI was adopted for use as a DNA probe due to its efficacy in binding DNA and its large subsequent increase in fluorescence. DAPI binds particularly strongly to A-T rich regions of double-stranded DNA and will experience a roughly 20-fold increase in fluorescence upon binding. It can also bind to RNA, however the fluorescence increase is weaker and the emission is red-shifted.
DAPI is commonly used for fluorescence microscopy. It is particularly successful at staining dead cells or cells with compromised membranes. DAPI can pass through intact cell membranes, albeit with difficulty. Thus, it is not recommended for live cell staining. DAPI has an excitation of 358 nm and an emission of 461 nm. This means it will appear blue when visualized with a blue/cyan filter. It can also be used in conjunction with green-fluorescent probes such as GFP. In experiments, DAPI has high cytotoxicity, which reinforces the reason to avoid using DAPI for live cell staining. DAPI is fairly non-toxic to humans if exposure occurs. However, as with many of the DNA probes, it is possible for DAPI to have some mutagenic properties.
PicoGreen®/Helixyte™
Pros & Cons
+ Extremely sensitive
+ Suitable for multiple applications
- Expensive
Applications
DNA quantification, PCR, microarrays
PicoGreen (and its chemical equivalent Helixyte) is an extremely sensitive probe for double stranded DNA. It can be used for biochemical applications such as quantification of dsDNA in a microplate reader. It can detect as little as 25 ng/mL of dsDNA. This makes it much more sensitive than the Hoechst stains. In addition to its sensitivity, PicoGreen is a very selective probe. It has high affinity for dsDNA over single stranded DNA, RNA and free nucleotides.
PicoGreen, as the name suggests, emits a green fluorescence when bound to DNA. It will also experience a roughly 1000-fold increase in fluorescence. This means it will have a very good signal to noise ratio. It can be used in a variety of applications, such as PCR and micro-array assays. It is a good replacement for DAPI and Hoechst. In particular, it good for when DNA samples are available in limited quantities, thanks to its greater sensitive and selectivity over traditional stains. These advantages come at a price, however. The probe itself is significantly more expensive than traditional stains.
| Cat-No. | Item | Size | Price (CHF) |
|---|---|---|---|
| 17591 | Helixyte Green *20X Aqueous PCR Solution* | 5 x 1 ml | 299.00 |
YOYO®-1/DiYO™-1/TOTO®-1/DiTO™-1
Pros & Cons
+ Extremely sensitive
- Cell-impermeable
- Expensive
Applications
Flow cytometry, cell viability assays
These are a family of cyanine dyes based around the compound oxazole yellow. Despite the name however, these dyes actually fluoresce green rather than yellow. For example, YOYO-1 (and its chemical equivalent, DiYO-1) has a maximum emission of 509 nm. The same is true of TOTO-1 (and its chemical equivalent, DiTO-1); this compound has a maximum emission of 535 nm. These compounds are intercalating agents, meaning they will insert themselves between the planar surfaces of DNA base pairs.
This family of DNA probes is particularly well known for its high affinity for DNA. Upon binding to DNA, these probes can experience a one thousand to three thousand fold increase in fluorescence. This strong affinity for DNA makes them excellent for experiments which require great sensitivity. The trade-off though is that these probes are fairly expensive.
Because these probes are cell-impermeable, they cannot pass through intact cell membranes. They are well suited for staining fixed or dead cells. In this regard, these probes have been commonly used in cell viability and cytotoxicity assays in conjunction with platforms such as flow cytometry.
| Cat-No. | Item | Size | Price (CHF) |
|---|---|---|---|
| 17575 | DiTO-1 [equivalent to TOTO-1] *5 mM DMSO Solution* *CAS 143413-84-7* | 0.2 ml | 299.00 |
| 17576 | DiTO-3 [equivalent to TOTO-3] *5 mM DMSO Solution* | 0.2 ml | 299.00 |
| 17580 | DiYO-1 [equivalent to YOYO-1] *5 mM DMSO Solution* | 0.2 ml | 299.00 |
| 17581 | DiYO-3 [equivalent to YOYO-3] *5 mM DMSO Solution* *CAS 156312-20-8* | 0.2 ml | 299.00 |