Scientists purchase antibodies with the expectation that they will recognize their intended targets and produce reliable data, but the presence of improperly validated antibodies on the market can make this process both time-consuming and expensive. Target recognition and specificity is fundamental to ensuring the rigor and reproducibility of data generated by immunoblotting, immunohistochemistry, ELISA, and other assays that rely on antibodies.
Bethyl antibodies are highly regarded for passing strict validation testing before arriving in customers’ hands. The principles that have guided our rigorous validation practices for decades preceded a recent publication* that describes five conceptual pillars for validating antibodies in an application- and context-specific manner.
* Uhlen M, Bandrowski A, Carr S, Edwards A, Ellenberg J, Lundberg E, Rimm DL, Rodriguez H, Hiltke T, Snyder M, Yamamoto T. A proposal for validation of antibodies. Nat Methods. 2016 Oct;13(10):823-7. PMID: 27595404.

We believe our customers can make a more confident purchase when they know how Bethyl antibodies are validated. Below are the six pillars we apply in the lab every day for antibody validation. Multiple pillars are used in a complementary fashion to validate each antibody, and the pillars can be variable based on the unique biology of each target protein, or the reagents available for that project.
Click each pillar for a detailed description and an example dataset:
- Pillar 1: Independent Antibodies
This pillar requires that two or more antibodies directed against different epitopes of a protein generate similar results.
- Pillar 2: Complementary Assays
This pillar requires that multiple, antibody-dependent assays produce complementary results.
- Pillar 3: Orthogonal Characteristics
This pillar requires that antibody-independent and antibody-dependent assays produce results that are correlative.
- Pillar 4: Biological Characteristics
This pillar takes advantage of the unique biology associated with some protein targets.
- Pillar 5: Protein OE/Epitope Tags
This pillar uses over-expressed (OE) proteins to validate antibodies against targets where we cannot identify a natively expressing cell line, or the protein is expressed at levels insufficient for detection.
- Pillar 6: Genetic Strategies
This pillar uses gene knockout or knockdown to reduce the levels of target protein available for detection.
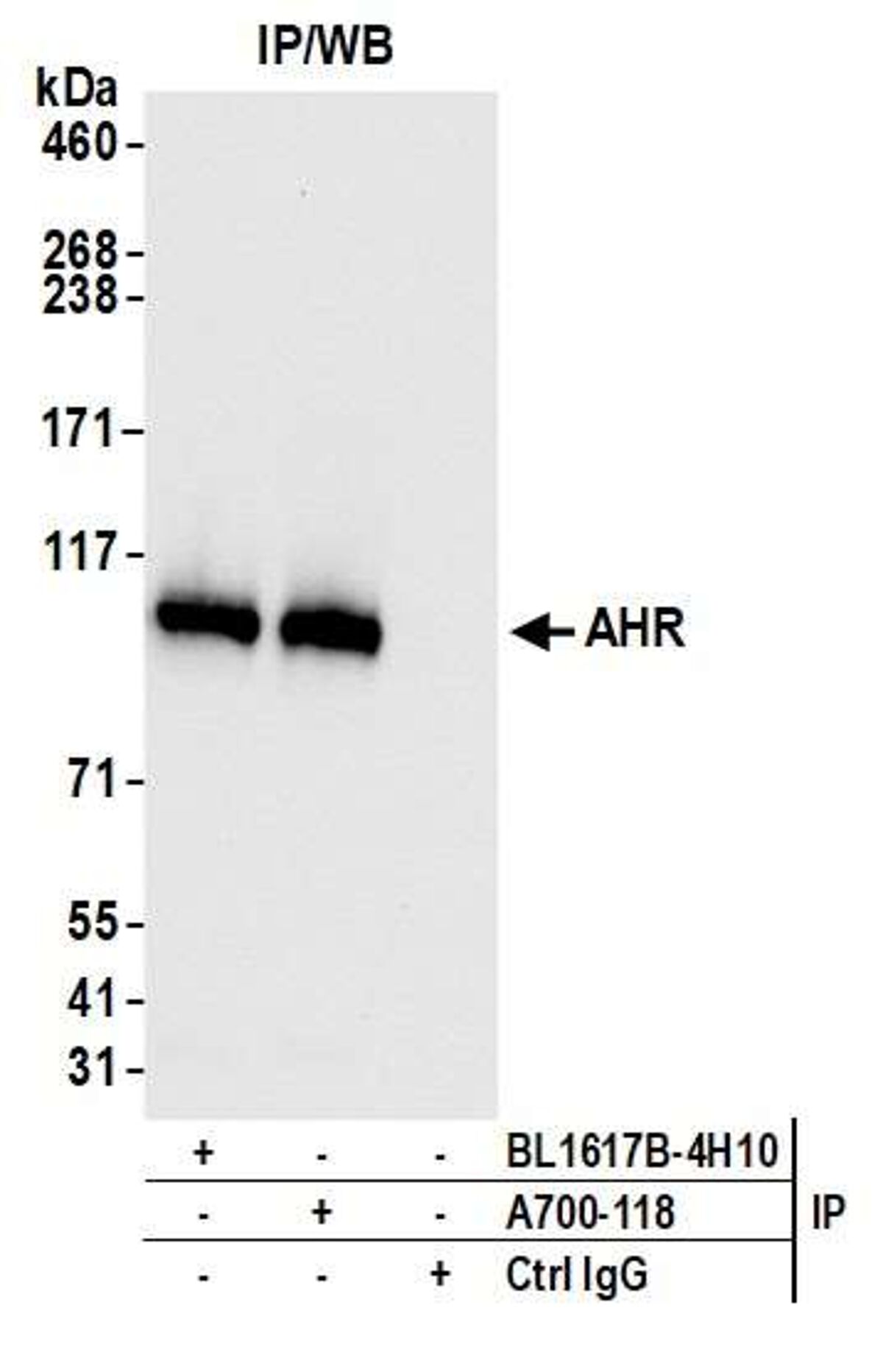
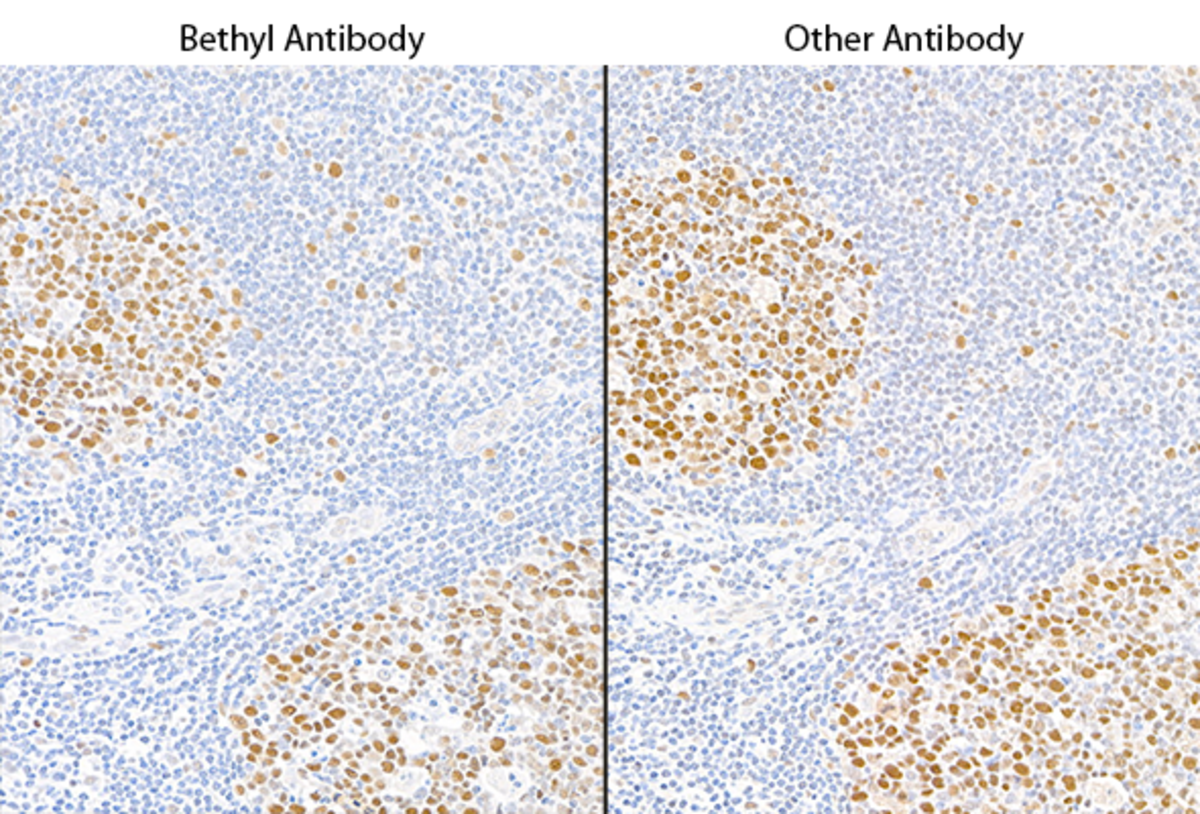
Pillar 1: Independent Antibodies
This pillar requires that two or more antibodies directed against different epitopes of a protein generate similar results. For example, two antibodies targeting different epitopes on a given protein target should produce similar staining patterns on the same tissue in an immunohistochemistry (IHC) study or produce similar results on a western blot. The majority of Bethyl antibodies are validated using independent antibodies developed in-house.
Example of data for this pillar:
Immunohistochemistry (IHC): We used two antibodies against MSH6 (Bethyl antibody A700-117 and a competitor) to stain serial sections of a tissue. Both antibodies generated a similar staining pattern.
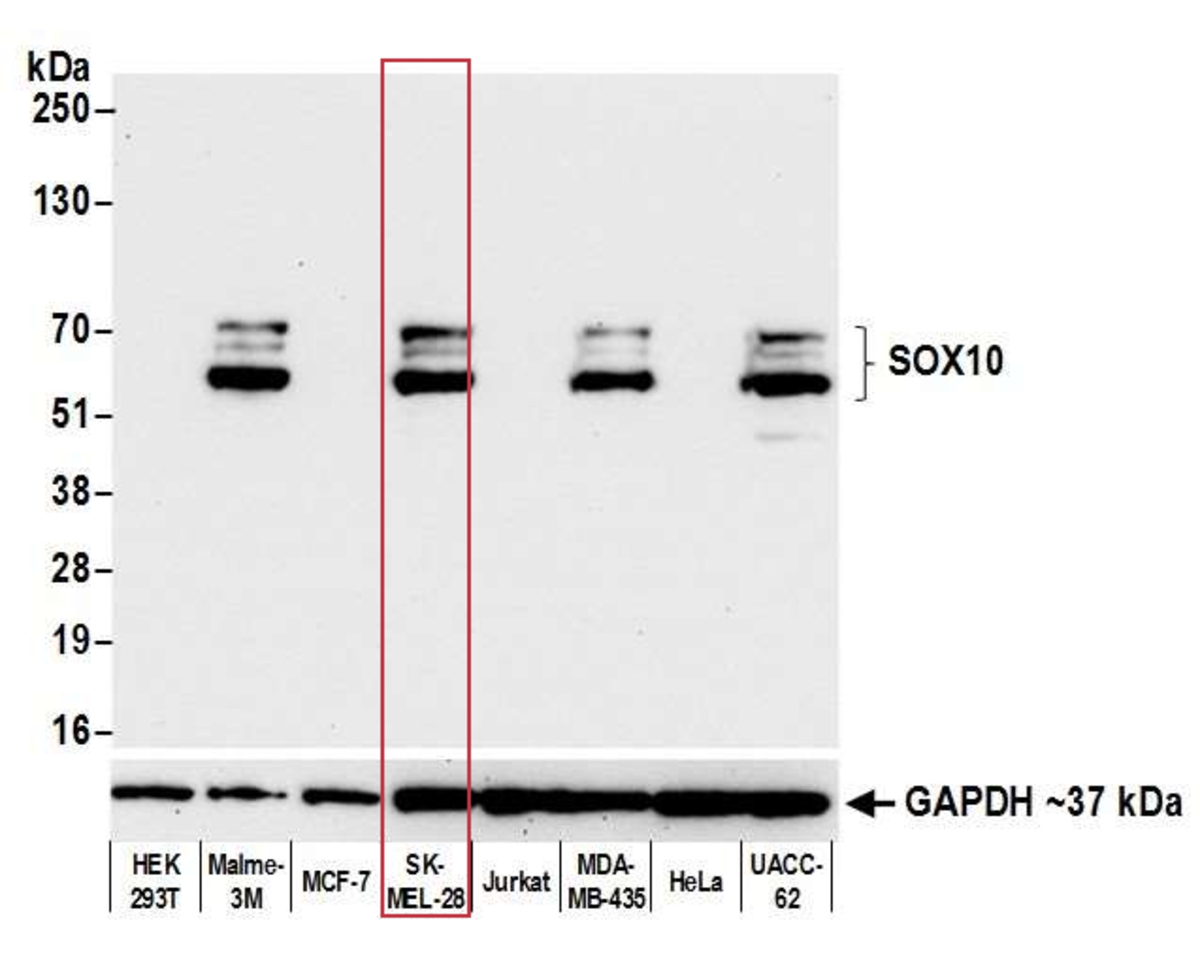
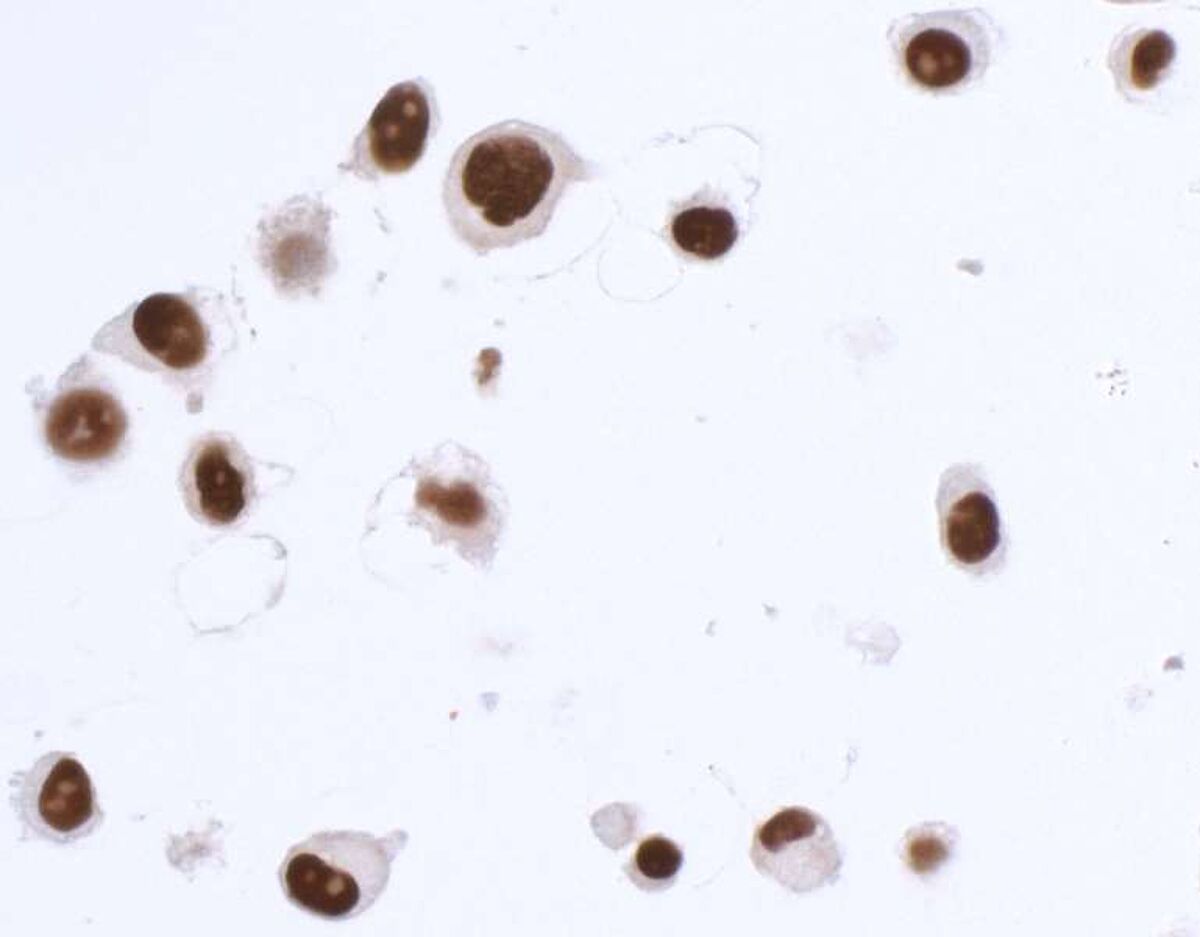
Pillar 2: Complementary Assays
This pillar requires that multiple, antibody-dependent assays produce complementary results. For example, when the same antibody of interest produces cell line-specific staining in an immunocytochemistry (ICC) assay and shows similar specificity on a western blot. Additional antibody-dependent assays under this pillar include immunoprecipitation (IP), immunohistochemistry (IHC), flow cytometry, chromatin immunoprecipitation (ChIP), and competitive ELISA. This pillar also captures the second half of a reciprocal IP experiment—for example, an IP with an independent antibody (Pillar 1), followed by western blotting with the antibody of interest.
Example of data for this pillar:
The SK-MEL-28 cell line expresses SOX10 protein. We used Bethyl antibody A700-080 to detect the SOX10 protein in cell lysate by western blot analysis (left). We used the same antibody to detect SOX10 by ICC (Right). The antibody produced complementary results in both experiments.
Pillar 3: Orthogonal Characteristics
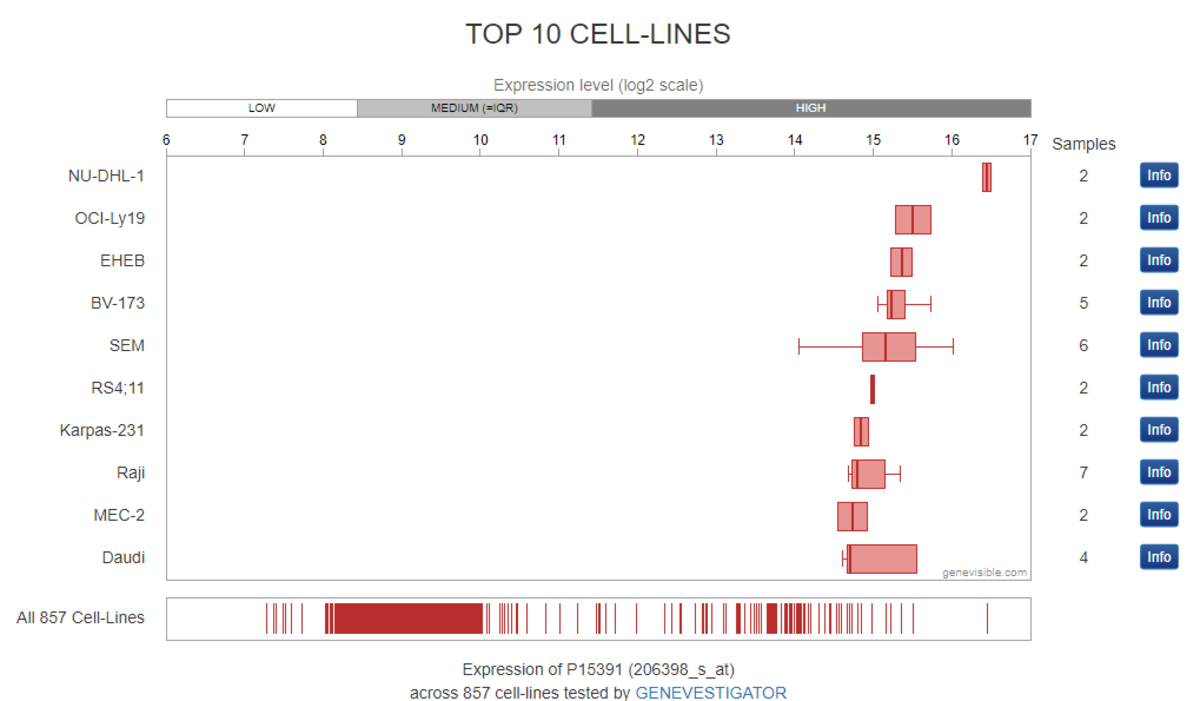
This pillar requires that antibody-independent and antibody-dependent assays produce results that are correlative. The antibody-independent techniques typically involve mass spectroscopy or mRNA transcript analysis of cell lines and tissue samples. We retrieve this data using both proprietary databases and public databases, such as GeneVisible or ProteomicsDB. We look for a clear relationship between these orthogonal data and western blot or immunocytochemistry (ICC) data generated using our antibodies in specific cell lines.
Example of data for this pillar:
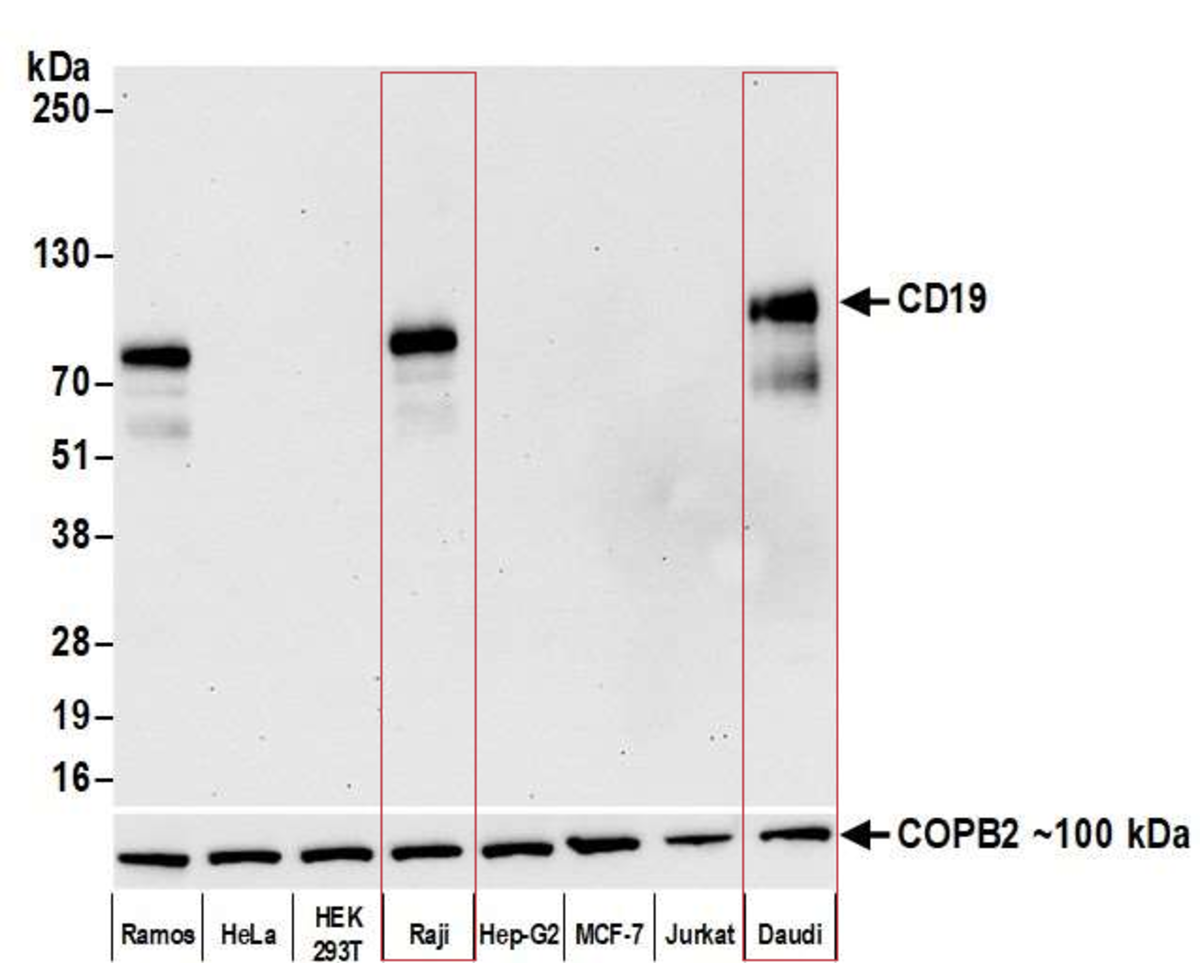
We used a public mRNA expression database (GeneVisible) to identify both Raji and Daudi cell lines as expressing the protein CD19 (this data is not provided). The corresponding western blot using our antibody of interest confirms the presence of CD19 (Bethyl antibody A700-137) in Raji and Daudi cell lysates.
Pillar 4: Biological Characteristics
This pillar takes advantage of the unique biology associated with some protein targets. To satisfy this pillar, the antibody of interest detects a protein target (or post-translational modification) that is only expressed or present under specific conditions (e.g., hypoxia, drug treatment). A phosphatase treatment on a phosphorylated protein can also be utilized to show phospho-specificity. We commonly use this pillar to validate phospho-specific antibodies.
Example of data for this pillar:
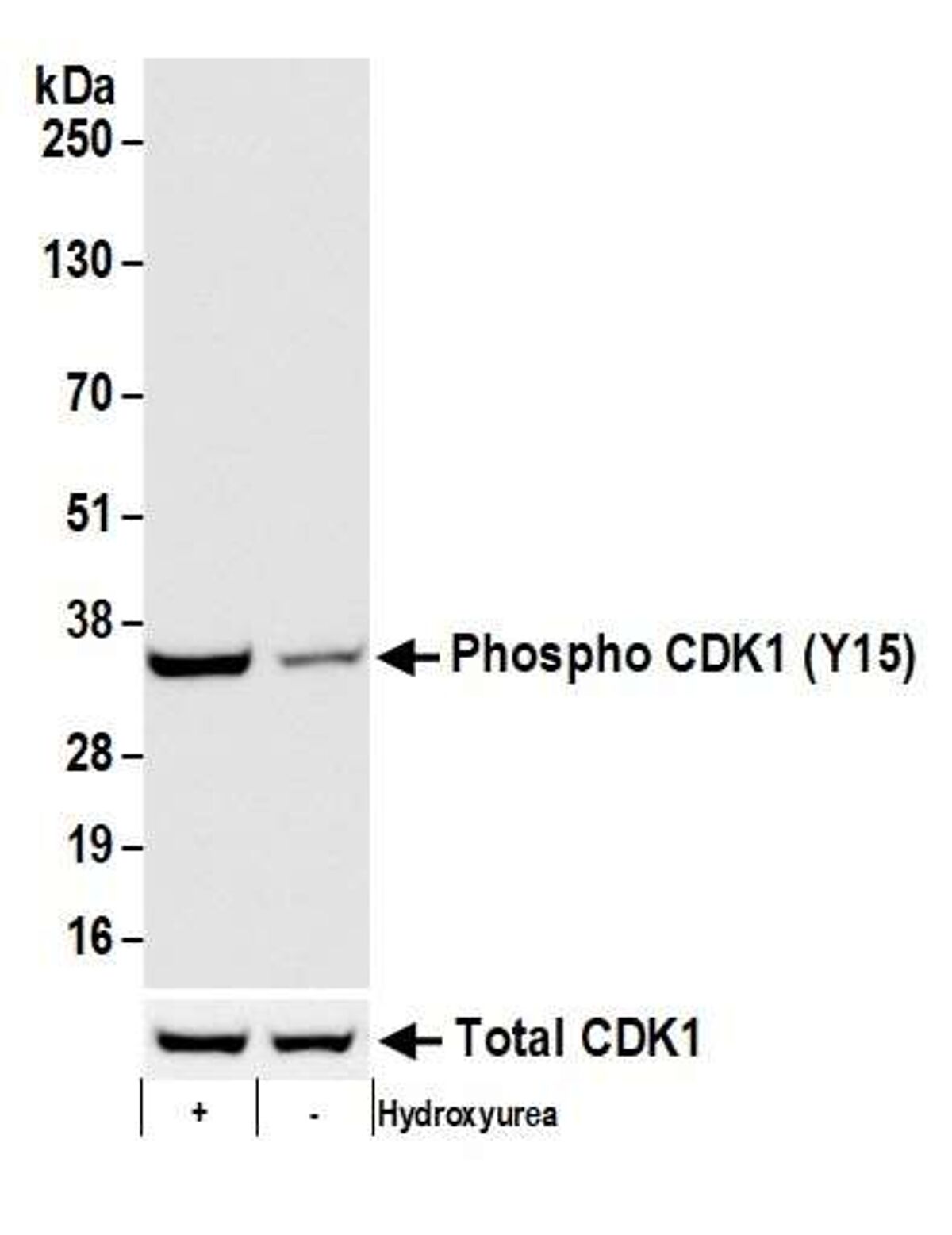
In this example, our antibody is specific for a phosphorylated version of CDK1 protein (Bethyl antibody A700-101), which is induced by the addition hydroxyurea. In this western blot, our antibody of interest detected the increased phosphorylation of CDK1 at tyrosine 15 when cells were treated with hydroxyurea.
Pillar 5: Protein OE/Epitope Tags
This pillar uses over-expressed (OE) proteins to validate antibodies against targets where we cannot identify a natively expressing cell line, or the protein is expressed at levels insufficient for detection. We also use this pillar to rule out cross-reactivity with specific proteins. This pillar is always combined with one or more pillars.
Example of data for this pillar:
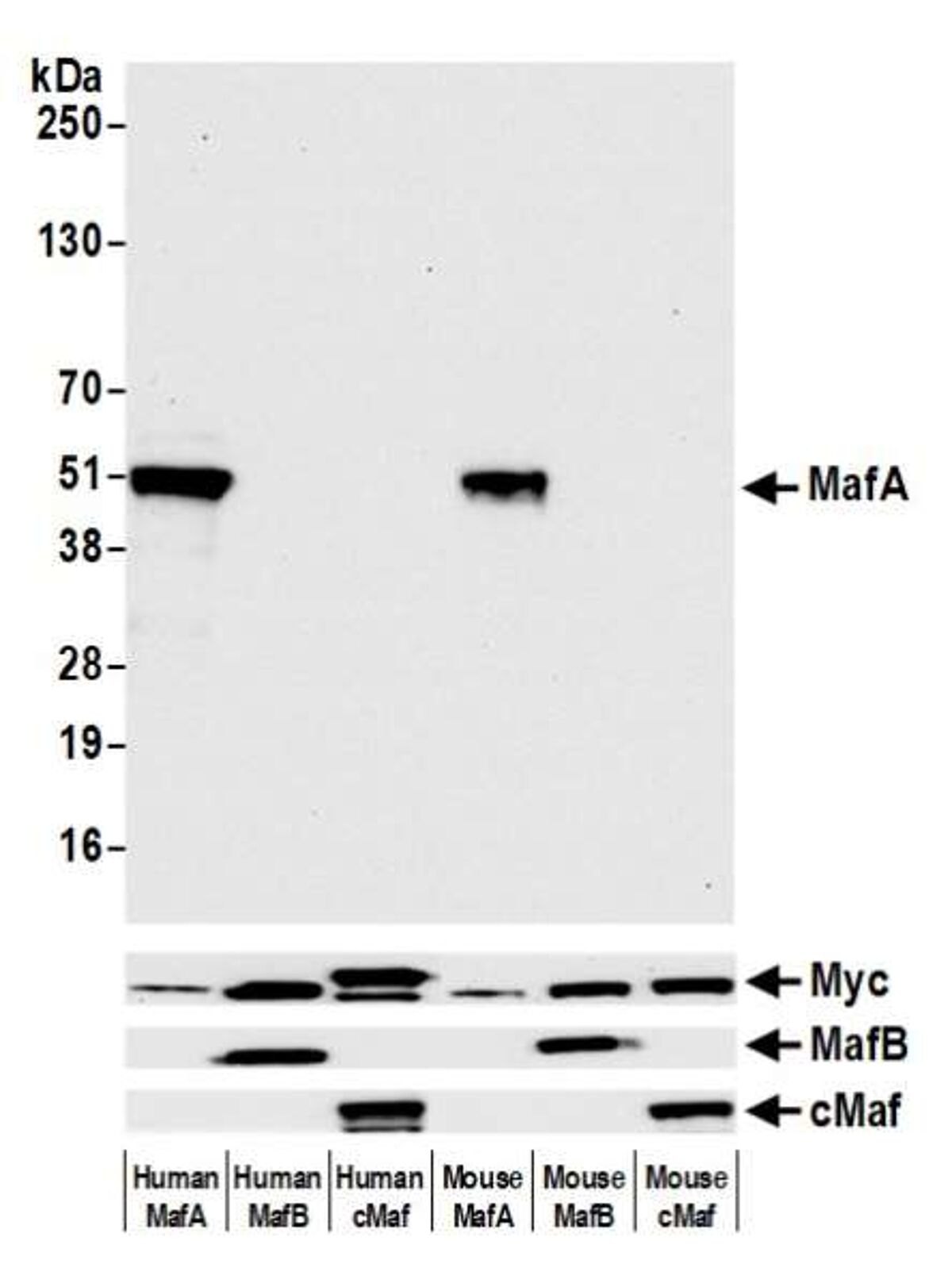
In this example of testing for cross-reactivity, we used mouse and human OE cell lines expressing MafA, MafB, and cMaf proteins. We then performed western blot of the cell lysates with Rabbit anti-MafA recombinant monoclonal antibody (A700-067) to show that our MafA antibody binds only to MafA, but not MafB or cMaf. The control bands show that the other proteins are present but are not being detected by the MafA antibody.
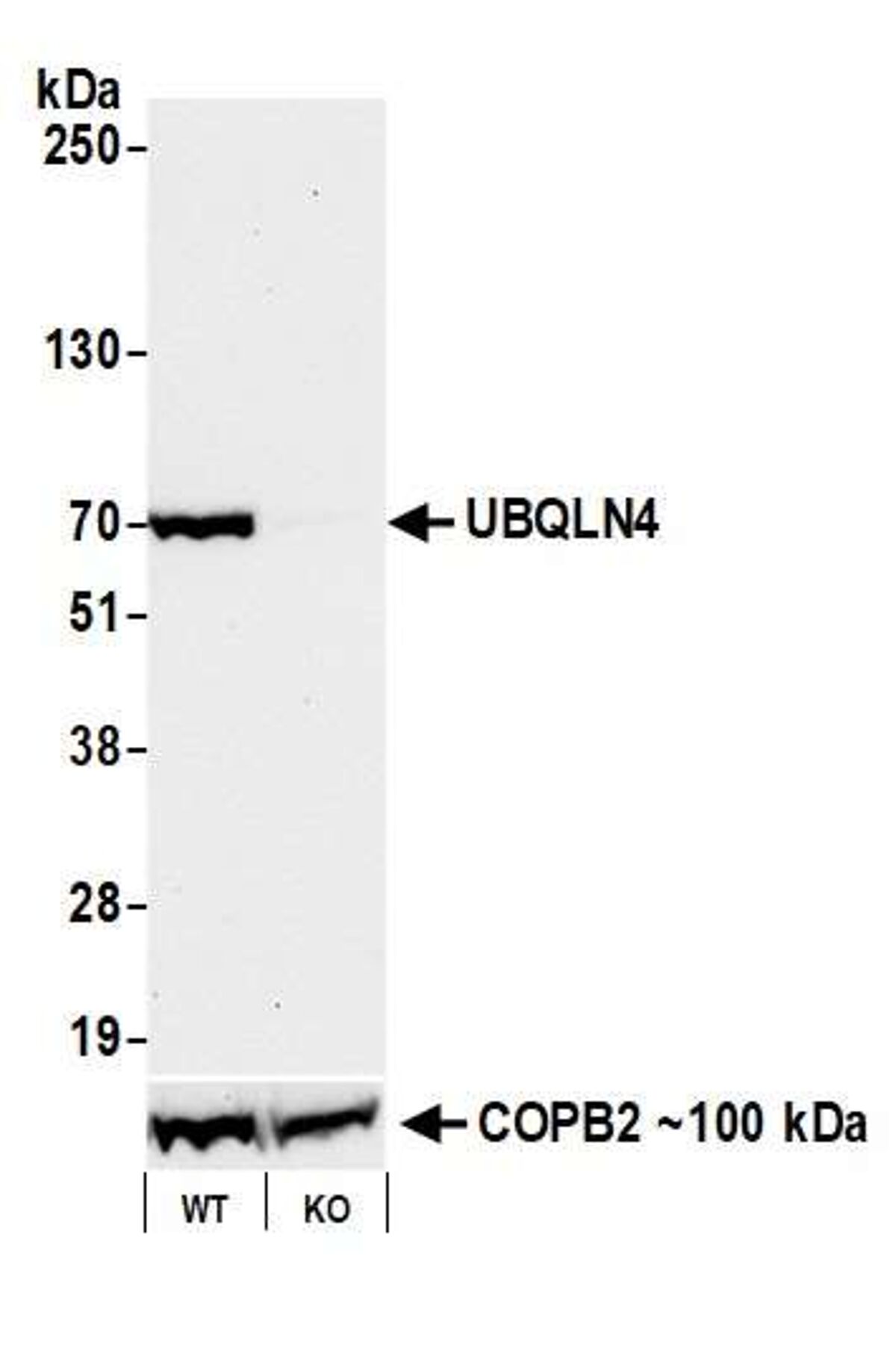
Pillar 6: Genetic Strategies
This pillar uses gene knockout or knockdown to reduce the levels of target protein available for detection. For example, a knockout of a target protein should ideally result in a reduced band intensity on a western blot, or an abrogation of signal in those cells in an immunocytochemistry (ICC) experiment. This is not a commonly used pillar for Bethyl antibodies.
Example of data for this pillar:
In this example, we use our antibody to UBQLN4 (Bethyl antibody A700-145) protein by western blot. Target protein is detected in lysates prepared from wild type cells, but not with lysates from the knock-out (KO) cell line. This provides a direct link between the gene, the target protein, and our antibody.
Supplier

Bethyl Laboratories - Fortis Life Sciences applies B-cell sorting and recombinant DNA technology to deliver high quality recombinant rabbit monoclonal antibodies. The pillar strategy validation ensures that the antibody works in the stated applications.