Built for homology-directed repair experiments
For researchers looking to accelerate their CRISPR, HDR-mediated insertions (>120 bases), IDT's dsDNA templates offer a cost effective, high-fidelity option to reduce the amount of downstream screening via higher HDR rates and lower unintended integrations
- Ideal for making longer than 120 bp genomic insertions
- Sequence-verified via next-generation sequencing
- Modified to increase successful HDR events
- Lower unintended and non-homologous (blunt) integrations
HDR Donor Blocks include chemical modifications within universal, non-integrating terminal sequences to help reduce unwanted blunt integration events.
Alt-R™ HDR Donor Blocks offer an improved solution for efficient generation of large knock-ins compared to long, ssDNA templates
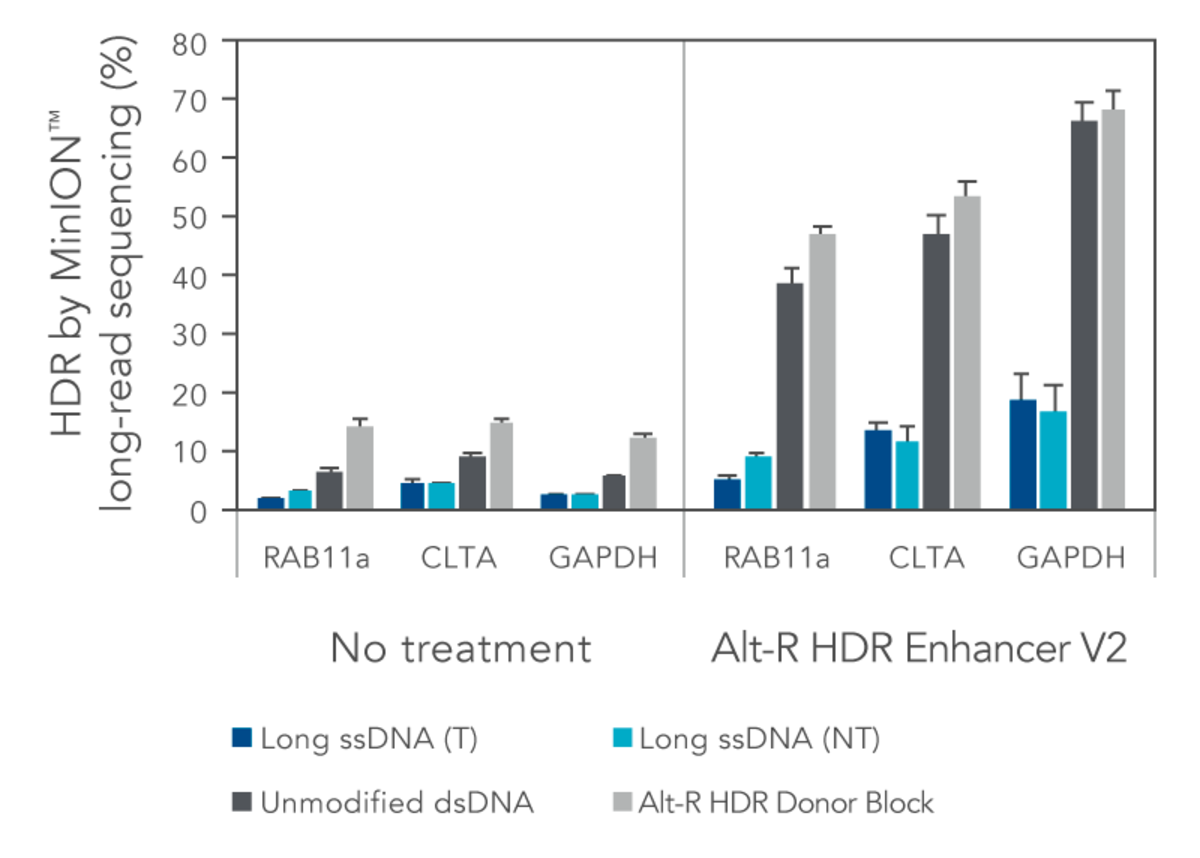
IDT investigated the HDR efficiency of inserting a green fluorescent protein (GFP) tag using either long, single-stranded DNA (ssDNA) or double-stranded DNA (dsDNA) templates (700 bp insert, 200 bp homology arms). The combined use of the modified Alt-R HDR Donor Blocks and the Alt-R HDR Enhancer V2 gave higher HDR rates compared to no treatment when tested at multiple genomic loci and in multiple cell lines (Figure 1).
Figure 1. HEK-293 and K562 cells were electroporated with 2 µM Cas9 RNP complexes targeting three genomic loci along with 50 nM dsDNA or ssDNA donor templates using the Nucleofector™ system (Lonza). HDR templates were designed to mediate a GFP-tagging event (700 bp insert, 200 bp homology arms). The dsDNA templates contained either no modifications (unmodified), or the Alt-R HDR Donor Block modification. Both the targeting (T) and non-targeting (NT) strands were tested for the long ssDNA templates. After electroporation, cells were plated in media with or without 1 µM Alt-R HDR Enhancer V2 with a media change after 24 hours. Genomic DNA was isolated 48 hours after electroporation. Editing was assessed by long-read amplicon sequencing on the MinION™ system (Oxford Nanopore Technologies) using R9.4.1 chemistry and Guppy High Accuracy (HAC) basecalling. Reads were mapped to the unedited reference amplicon or desired HDR outcome using minimap2 (-x map-ont). A read was classified to be derived from the HDR pathway if it preferentially mapped to the desired HDR outcome. HDR efficiency was measured by calculating the read counts classified as HDR relative to the total aligned reads per sample. Error bars represent the standard error of two biological replicates per cell line.
Combined use of Alt-R HDR Donor Blocks and Alt-R HDR Enhancer V2 assist in mitigating the risk for off-target integration events
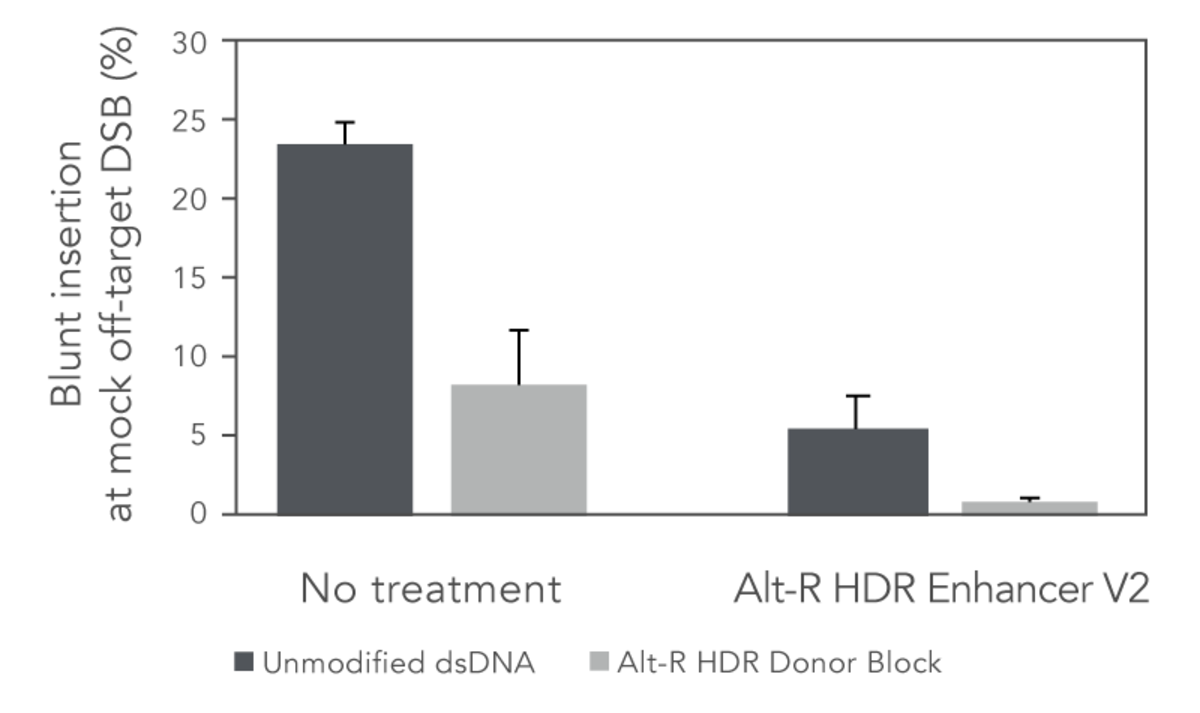
Since use of dsDNA templates may pose a risk for unwanted off-target integrations, the ability of the Alt-R HDR Donor Blocks to reduce the occurrence of this unwanted repair event was examined. As shown in Figure 2, use of Alt-R HDR Donor Blocks reduced the blunt insertions at a mock off-target double strand break by 65% compared to unmodified dsDNA. Addition of Alt-R HDR Enhancer V2 further reduced the off-target integration, lowering blunt insertion levels to 1% (Figure 2).
Figure 2. Combined use of Alt-R HDR Donor Blocks and Alt-R HDR Enhancer V2 reduces the rate of non-homologous (blunt) integrations at off-target DSBs. A mock off-target DSB was generated by delivering 2 µM Cas9 RNP targeting the SERPINC1 locus into HEK-293 cells using the Nucleofector™ system (Lonza). The dsDNA donor templates mediating GFP insertions at alternative genomic loci (50 nM dose, n=4 sequences) were co-delivered with the mock off-target RNP. After electroporation, cells were plated in media with or without 1 µM Alt R HDR Enhancer V2 with a media change after 24 hours. Genomic DNA was isolated 48 hours after electroporation. The SERPINC1 locus was PCR-amplified and blunt insertion events were measured by size discrimination on a Fragment Analyzer (Agilent).