The Importance of Microbiota
Don't look now, but you're outnumbered by a factor of more than 10 to 1. You might not know it, but your body is basically an apartment building for a wide variety of microbes. Your skin, respiratory system, gastrointestinal tract, and everything in between is home to a complex and unique-to-you ecosystem of fungi, bacteria, viruses, and protists. Fortunately, many of these tiny tenants are paying rent, either by helping fend off harmful invaders, assisting with the digestion of food, or a wide variety of other beneficial tasks. The human microbiota is a complex community made of diverse microbes that play an integral part in our health.1
A symbiotic relationship, beginning at childbirth, develops between the human body and its indigenous microbiota which play an important role in maintaining the health and general well-being of an individual.2 This relationship is constantly evolving in response to changes in your life, such as age, nutrition, lifestyle, hormonal changes, disease, and genetics.2 Balance in the composition of the microbiota is critical in maintaining the health of an individual, as shifts from this balance (dysbiosis) can lead to disease or even death in some cases.2
Even though the association of multiple diseases with dysbiosis has already been well-established, there is still a missing gap of knowledge on organism-specific metabolic activities that lead to disease.1 The only way to fill this gap is to analyze the gene expression of the gut microbiota in healthy individuals and compare it to that of someone with a disease of interest.
Sampling Methods for Gut Microbiota
Metatranscriptomics is the analysis of the expressed genes in the entire microbial community of a given habitat.3 In order to analyze the collective transcriptome (sum total of all messenger RNAs) of an entire microbial community inhabiting the gut, a convenient and easily accessible sample type is required. A critical step in this process is the collection of appropriate samples of the gastrointestinal microbiota. Commonly used collection methods include mucosal biopsy, intestinal aspiration, luminal brushing, and fecal collection.4 The easiest and most commonly used method to sample gut microbiota is through feces. It is convenient, non-invasive, inexpensive, and can be sampled repeatedly to provide a measure of change over time.
With recent advances in high throughput sequencing, it has become possible to sequence and analyze the vast transcriptome produced by the gut microbiota, known as the metatranscriptome. This method of sequencing provides researchers with an accurate method of analyzing the metagenome and metatranscriptome relatively in situ!4

The Link Between Dysbiosis and Colorectal Cancer
As mentioned previously, gut microbiota dysbiosis has been linked to diseases like colorectal cancer. Important to note here is that bacteria alone can not cause carcinogenesis; it is the host-microbiome interactions that lead to the development of cancer. These interactions are mediated by proteins, metabolites, and small RNAs (sRNAs), including Homo sapiens microRNAs (hsa-miRNAs) and other non-coding small RNAs (hsa-sncRNAs).5 Liu and colleagues (2016) observed that alteration of mouse intestinal miRNA synthesis promoted gut dysbiosis which could be restored by fecal transplantation of mouse miRNAs.6 In the same study the authors also observed that specific hsa-miRNAs can affect the growth of the bacterial cells by regulating gene expression.6
Bacterial small RNAs (bsRNAs) may also play a role in carcinogenesis, but their involvement in host-microbiome interactions has not yet been fully explored. Like hsa-miRNAs, bsRNAs may also play a role in regulating gene expression in both human and microbial cells. Even though bsRNAs can be detected in transcriptome experiments, their identification and quantification is difficult due to their small size. Not all is lost though, as integrating small RNA sequencing (sRNA-seq) with whole metagenomic sequencing profiles could make the identification of microbial sRNAs more reliable for finding potential colorectal cancer biomarkers. Using this approach, Tarallo and colleagues (2019) characterized the human and microbial sRNAs of stool samples from healthy individuals and patients with colorectal adenoma or carcinoma. Through this study, the authors identified a human and microbial sRNA signature that can be used to improve the diagnosis of colorectal cancer.
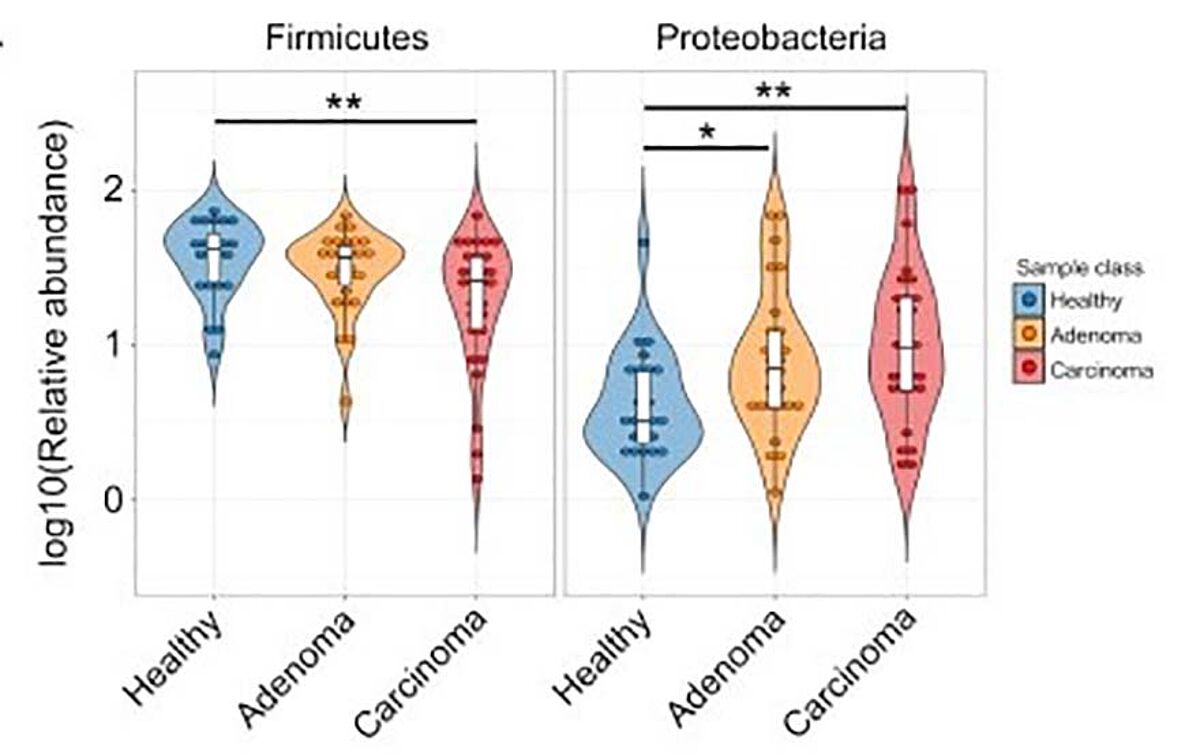
The end result of the experiment showed a strong profile for all three groups, healthy, adenoma positive, and carcinoma positive. On a bacterial phylum level, Firmicutes were the predominant group present in the carcinoma patients, with a significant increase in Proteobacteria as well (Figure 1).5 Stepping in for a closer look at the molecular level, 450 differentially expressed bsRNAs were discovered, with over a third of them being attributed to E.coli.5 Of note, several of these bsRNAs promote more pathogenic E. coli strains that are better suited for the nutrient-deprived, anaerobic conditions inside of tumors and adenomas. Moving to the human side of things, 43 hsa-miRNAs showed different expression patterns in those with CRC.5 These miRNAs can potentially be used as biomarkers for colorectal cancer. The study also identified 13 hsa-miRNAs that were significantly correlated with E. coli abundances (Figure 2).5 The authors hypothesized that these hsa-miRNAs interact with bacteria via their target genes to modulate bacterium adhesion and phagocytosis.
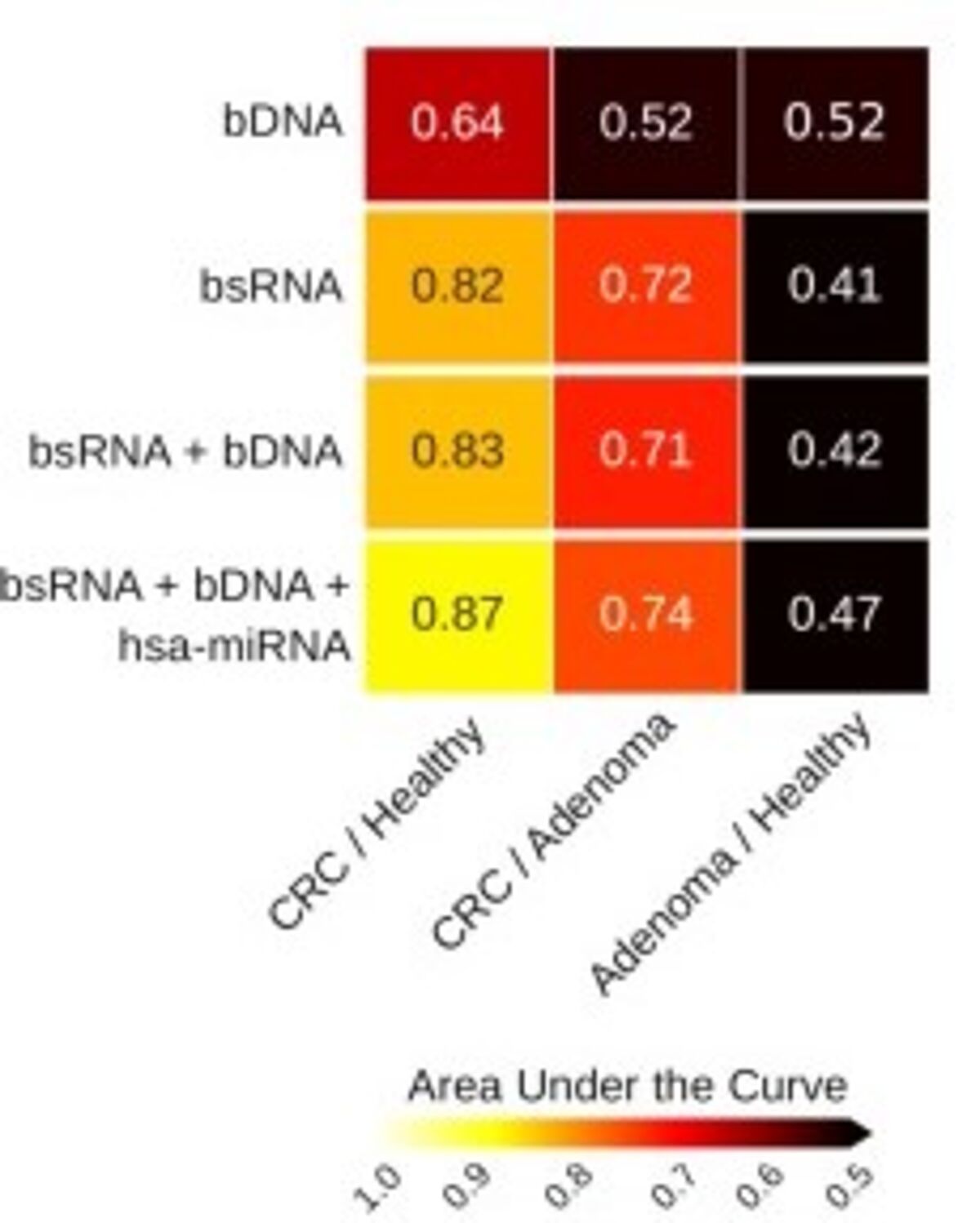
By further combining the metagenomic, metatranscriptomic, and transcriptomic profiles, using bacterial DNA (bsDNA), hsa-miRNA and bacterial small RNA (bsRNA), the authors were able to further classify CRC when compared to controls and adenomas (Figure 3).5
Understanding Microbial Composition
In conclusion, the metatranscriptomic analysis provides a powerful tool to not only identify the difference in stool microbial composition between healthy, adenoma, and CRC groups, but also the microbial species transcriptionally active in adenoma and CRC patients. Furthermore, the combined use of metatranscriptomic and transcriptomic data identified human and bacterial RNA biomarkers that can potentially be used to diagnose patients with colonic adenomas or carcinomas. Finally, metagenomic and metatranscriptomic analyses, in combination, can provide information that can accurately classify subjects according to their health status.
As previously mentioned, one of the greatest difficulties for earlier gut microbiome studies was the fragility of the microbiome itself. Once outside the body, the overall composition begins to change and shift. In the past, this meant freezing the sample as rapidly as possible to preserve the microbiome profile. However, this caused issues since freezer storage or cold transport is not always available. Chemical preservation is a popular alternative to freezing as it eliminates the need for freezing, and in the case of Norgen's Stool Nucleic Acid Collection and Preservation Tubes, maintains the stability of DNA and RNA in the sample for 2 years and 7 days respectively while largely eliminating odor.
Extracting nucleic acids from stool samples can be a challenge, and under non-optimal collection and shipping conditions, nucleases in stool remain active and degrade nucleic acids. As a result, a stool sample, when it reaches the lab, may largely consist of DNA and RNA fragments that may be as small as 200 bp or lower. This poses a challenge for studies that aim to extract total RNA, including microRNA and small fragmented RNA, from stool samples. Traditional silica-based methods can not capture RNA molecules shorter than 200 nt unless supplemented with carrier-RNA or organic extraction via hazardous compounds, such as phenol/chloroform, both of which affect downstream applications. Norgen’s proprietary silicon carbide technology allows for the capture of true total RNA, including miRNAs and any fragmented RNAs present in the sample, without using any special reagents that adversely affect downstream applications.

If you are interested in learning more about Norgen’s Stool Nucleic Acid Collection and Preservation Devices and proprietary Silicon Carbide technology, be sure to watch their webinar, Get to the Bottom of Your Gut - A Multi-Omics Look Into the Stool Microbiome.
Sohaib Siddiqui | May 13, 2022
Supplier

Norgen Biotek
In addition to topselling Total RNA purification Kit line, Norgen also offers collection devices for whole blood (cf-RNA/DNA), urine, saliva and more.
References
Ranjan, R., Rani, A., Finn, P. W., & Perkins, D. L. (2018). Multiomic strategies reveal diversity and important functional aspects of human gut microbiome. BioMed Research International, 2018, 1–13. https://doi.org/10.1155/2018/6074918
Ogunrinola, G. A., Oyewale, J. O., Oshamika, O. O., & Olasehinde, G. I. (2020). The human microbiome and its impacts on health International Journal of Microbiology, 2020, 1–7.2.https://doi.org/10.1155/2020/8045646
Dubey, R. K., Tripathi, V., Prabha, R., Chaurasia, R., Singh, D. P., Rao, C. S., El-Keblawy, A., & Abhilash, P. C. (2019). Metatranscriptomics and metaproteomics for microbial communities profiling. Unravelling the Soil Microbiome , 51–60. https://doi.org/10.1007/978-3-030-15516-2_5
Tang, Q., Jin, G., Wang, G., Liu, T., Liu, X., Wang, B., & Cao, H. (2020). Current sampling methods for gut microbiota: A call for more precise devices. Frontiers in Cellular and Infection Microbiology, 10. https://doi.org/10.3389/fcimb.2020.00151
Tarallo, S., Ferrero, G., Gallo, G., Francavilla, A., Clerico, G., Realis Luc, A., Manghi, P., Thomas, A. M., Vineis, P., Segata, N., Pardini, B., Naccarati, A., & Cordero, F. (2019). Altered fecal small RNA profiles in colorectal cancer reflect gut microbiome composition in stool samples. MSystems, 4(5). https://doi.org/10.1128/msystems.00289-19
Liu, S., da Cunha, A. P., Rezende, R. M., Cialic, R., Wei, Z., Bry, L., Comstock, L. E., Gandhi, R., & Weiner, H. L. (2016). The host shapes the gut microbiota via fecal MicroRNA. Cell Host & Microbe, 19(1), 32–43. 6.https://doi.org/10.1016/j.chom.2015.12.005