What is in situ hybridization and what are the recent advancements with this indispensable technique?
Summary
- In situ hybridization (ISH) is a widely used method to localize and quantify DNA or RNA in a variety of cytological and histological preparations, preserving the morphology of cell and tissue.
- ISH is an extremely flexible technique. It is, in fact, possible to choose between DNA or RNA probes, which can bear either radio-, fluorescent- or antigen-labeled bases, allowing for a chromogenic (CISH) or fluorescence detection (FISH).
- Recent advancements in the technology allow to overcome its major limitation: the sensitivity, especially in the detection of single-copy targets. In this context, Enzo’s AMPIVIEW™ RNA probes, powered by Enzo’s LoopRNA ISH™ technology, are amongst the most sensitive means of detection of nucleic acid targets in tissue and cells, without damaging the morphology of the sample.
Introduction
In situ hybridization (ISH) is a widely used method to localize and quantify DNA or RNA in cytological preparations (metaphase spreads, cells, tissues), or even in whole mount embryos (WMISH). Its official birth dates back to 1969, when two independent research groups used radiolabeled RNA produced in vivo, as a probe to detect the complementary DNA in the nucleus of Xenopus oocytes (1,2). The radiolabeling technologies used for the first-generation probes looked promising, however, their application has been rather limited because of numerous constraints, such as the inherently short half-life of the isotopes employed for the labeling or, more importantly, the safety and environmental concerns related to radioactivity. Since the time of these first pioneering works, extraordinary advancements in molecular biology techniques made ISH more and more practical and versatile.

Today, ISH is a key laboratory tool, routinely used in a variety of fields with many different applications, from fundamental research to clinical diagnostic and prognostic. To achieve this flexibility, one can choose between DNA or RNA probes, which can bear either fluorescent- or antigen-labeled nucleotides. The detection method, chromogenic (CISH) or fluorescence (FISH), can also be adapted to the required type of analysis, with CISH being more suitable in molecular pathology diagnostics to visualize the target and specimen morphology simultaneously, and FISH being typically preferred for multiplexing and genomic aberrations studies.
This review will provide an overview of ISH and its basic principles. The newest advancements in the technique will also be introduced, describing how these progresses can further increase the range of the applications of ISH in the personalized medicine era.
How does ISH work?
Regardless of the specific protocol, which strictly depends on the sample type, on the aim of the study, as well as on the nature of the probe and on the chosen detection method, ISH procedure can be summarized in few key points:
- Probe generation: The probes, used as a sort of bait during the hybridization phase, are DNA or RNA fragments complementary to the sequence of interest. During the synthesis they can be labeled by the incorporation of nucleotides conjugated with a marker.
- Specimen preparation: The cells or tissue of interest are fixed for preservation. Tissue are typically embedded in paraffin and cut in thin sections. The specimen is then immobilized onto a glass slide and permeabilized to make the target sequence accessible to the probe.
- Denaturation: Both the probes and the nucleic acids in the samples are denatured by heat to make way for the subsequent hybridization phase.
- Hybridization (or annealing): It consists in the spontaneous pairing of the probe to its complementary DNA/RNA sequence in the sample.
- Detection: The hybrids formed between the probes and their targets can then be visualized exploiting the different types of labeling.
As described below, different probe types and detection methods are required depending on the goal of the study.
Type of probes
The hybridization can take place between complementary deoxyribonucleotides or ribonucleotides, therefore either DNA- or RNA-based probes can be used to localize DNA or RNA in a given sample. However, these different types of probes are not equivalent and one has to select the most suitable option for the experiment. For example, it is worth considering that RNA-RNA hybrids are more stable than RNA-DNA hybrids, which are in turn more stable than DNA-DNA hybrids. Depending on the ISH target, this difference in the stability has an influence on the choice of the probe nature.
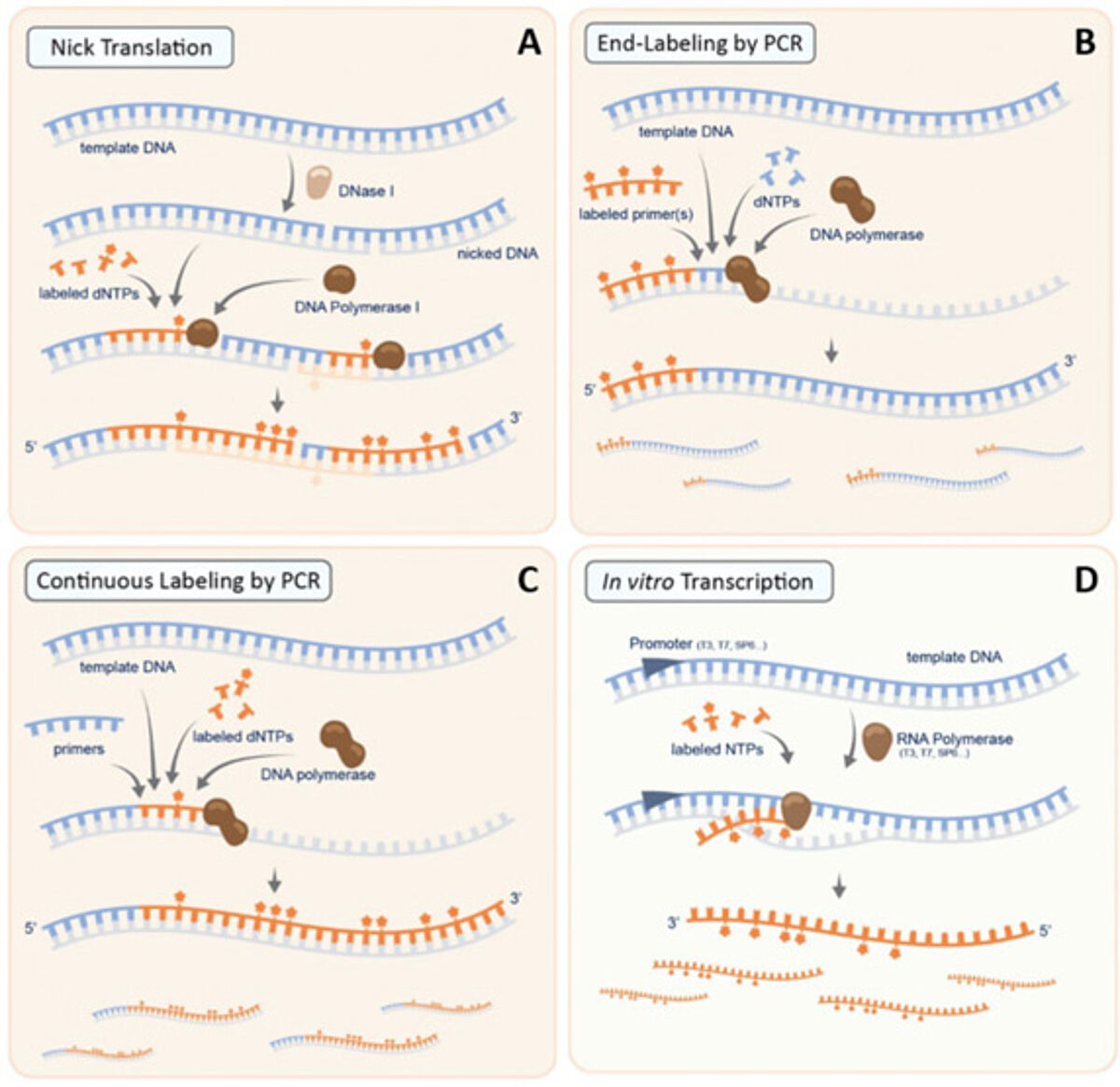
A DNA probe is a fragment of DNA that contains a nucleotide sequence specific for the chromosomal region of interest. One of the most common methods for their synthesis is the Nick translation reaction, occurring in two steps (Fig. 1A). First, the template DNA (usually a bacterial artificial chromosome [BAC]) is treated with low concentrations of DNAse I enzyme, which will nick the DNA randomly, producing a free 3′-OH group. The latter will act as a primer in the second step, when the DNA polymerase I will remove nucleotides by 5’-3’ exonuclease activity, while elongating the 3’-OH group. The modified nucleotides available in the reaction mix will then be incorporated in the newly synthetized strand, allowing the labeling of the probe. Polymerase chain reaction (PCR) is another common method for DNA probes synthesis: the amplification of the sequence of interest can be realized in the presence of appropriately labeled primers (End-labeling PCR, Fig. 1B) or, in alternative, the reaction mix can contain labeled dNTPs, so that the products of the reaction are labeled as they are being synthesized (Continuous-Labeling PCR, Fig. 1C). DNA-based ISH is more commonly used to detect specific DNA sequences in the nuclei, identifying for example potential gene amplifications, deletions, translocations, and chromosomal number.
RNA probes are stretches of single-stranded RNA used to detect the presence of complementary nucleic acid sequences by hybridization. RNA probes can be efficiently and consistently generated by in vitro transcription. A common strategy to do so is to clone the DNA to be transcribed between two promoters in opposite orientations, in order to synthesize either the sense or the antisense RNA using the appropriate polymerase. In this case, the RNA probes are continuously labeled as they are being transcribed, via the incorporation of modified nucleotides, thereby generating probes with a high degree of label incorporation (Fig. 1D). In addition, since RNA probes are synthesized from a linearized template, all probe molecules are of uniform length. RNA probes offer several key advantages associated with their use, including an improved signal with respect to DNA probes. However, because of the intrinsically labile nature of RNA and the susceptibility to RNase degradation, RNA probes must be treated with the same care as any other RNA preparations. RNA ISH is mainly used in the study of gene transcription, and thus of gene expression, at the cellular level.
Labeling type and detection method
As previously mentioned, ISH probes are labeled with nucleotides modified either with a fluorophore or an antigen. This difference is linked to the detection method.
Fluorophore-bearing probes allow for a direct fluorescence detection in fluorescence in situ hybridization (FISH). FISH is a highly sensitive technique, allowing the use of different fluorophores to visualize multiple target sites on the same specimen. FISH is useful for chromosome mapping and enumeration, as well as for the detection of structural rearrangements such as translocations, inversions, insertions and deletions. However, FISH can be limiting when information on tissue morphology is needed, thus a chromogenic detection may then be preferred.
CISH (Chromogenic in situ hybridization) is typically associated with an indirect detection and the use of biotin- or digoxigenin labeled probes.
The vitamin Biotin has been the first replacement to radiolabeled nucleotides, as it was possible to exploit its high affinity binding to avidin or streptavidin, in turn conjugated to a reporter enzyme [alkaline phosphatase (AP) or horseradish peroxidase (HRP)] for color development. Biotin can also be recognized by a specific antibody, with a downstream detection system pretty much identical to IHC/IF. However, biotin probe labeling can cause staining issues, in particular in tissues endogenously expressing this vitamin in high quantity, such as liver, kidney, skeletal and cardiac muscle, etc.
In response to this problem, digoxigenin-conjugated probes represent a valid alternative. Digoxigenin is a steroid antigen derived from plants of the genus Digitalis (3). Since it is found exclusively in these plants, it circumvents the possible background issue associated with biotin. The detection occurs via the specific binding with an anti-digoxigenin antibody. In addition, digoxigenin and biotin labeling can be combined in multiplexing, for instance, two different in situ probes can be used simultaneously, or if chromogenic ISH is combined with IHC.
Advancements in RNA ISH for single molecule visualization
Early diagnosis and personalized treatments are two of the major challenges that modern medicine faces. For this reason, the quantitative and qualitative analysis of DNA, RNA, and protein biomarkers is becoming more and more prominent in clinical practice, providing crucial information in diagnosis, prognosis, and therapy guidance (4). While DNA-based ISH and IHC are commonly applied in this context, the use of RNA-based ISH is still restricted to highly expressed genes (e.g. EBER1/2) (4), the main reason of that being the sensitivity of the technique, especially when it comes to the detection of single copy targets. For example, by using “classic” RNA ISH, the detection of HPV in cervicovaginal smears can be easily missed, as infections are often caused by a single integration of the viral genome in a small number of cells within a specimen (5). Therefore, RT-PCR is generally considered the gold standard for gene expression analysis in clinical diagnostic (as well as in any research field), since it allows the detection of very limited amount of the sequence of interest. Nonetheless, differently from ISH, RT-PCR requires the homogenization of the sample for DNA/RNA extraction, and consequently it cannot provide any spatial information concerning gene expression.
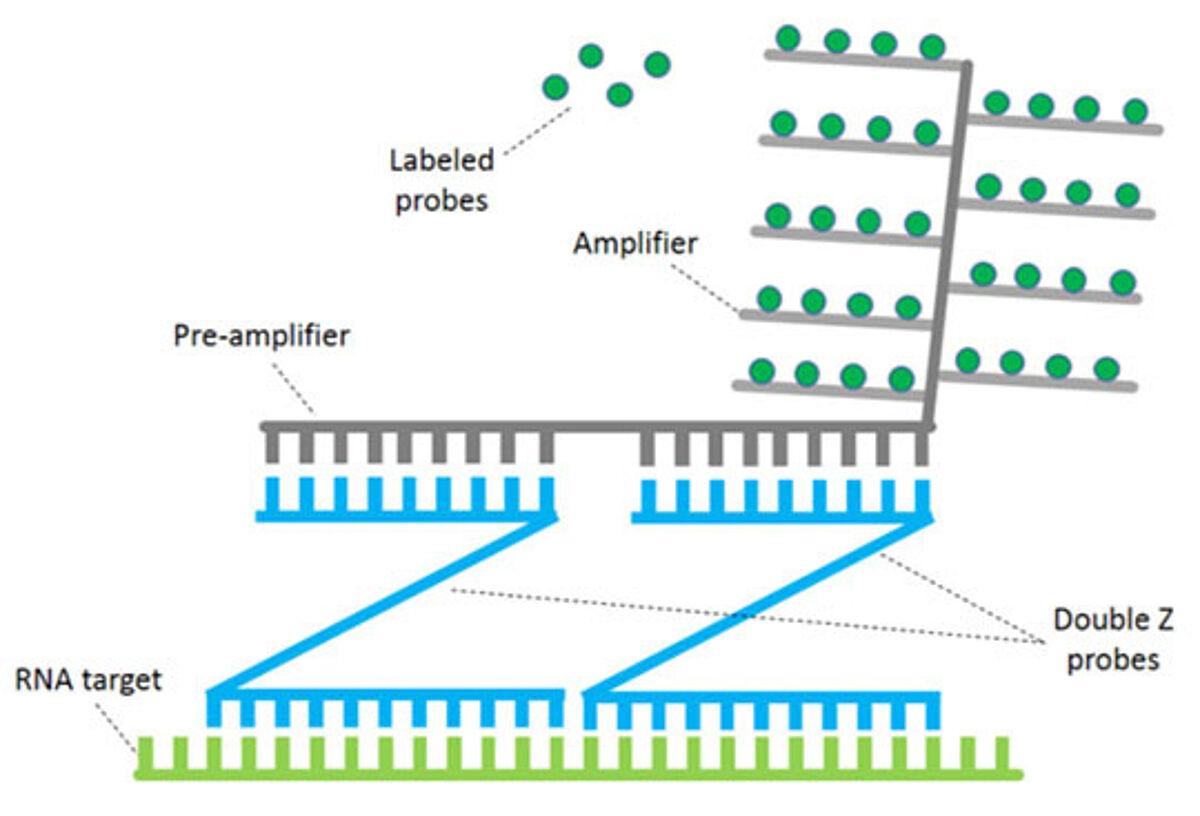
In order to overcome these limitations, continuous improvements on the ISH techniques have been introduced over the years, in an attempt to increase its sensitivity. The most common approach consists in enhancing the detection through the use of branched DNA (bDNA) (4,6), whose most popular application utilizes pairs of Z-probes (or double Z-probes). As illustrated in Fig. 2, a Z-probe is composed of three elements: a sequence complementary to the target RNA (necessary for the hybridization); a spacer (linker), and the Z-probe tail. The latter pairs with the pre-amplifier sequence, which in turn binds to multiple amplifiers. Finally, several labeled probes (chromogenic or fluorescent) attach to their specific sites on the amplifier molecules (7). The specificity of the system is assured by the fact that detection can occur only if both Z-probes are simultaneously annealed to the target, while the signal amplification process allows for an extremely elevated sensitivity.
Despite the numerous evident advantages of this form of bDNA technologies, some of its aspects can still constitute a constraint. For instance, given the multiple steps required for the amplification, the protocol is considerably long. The hybridization conditions can also be harsh, thereby altering tissue or affecting the possibility to visualize other markers. Finally, its overall elevated cost makes it less accessible to smaller laboratories.
AMPIVIEW™ RNA probes
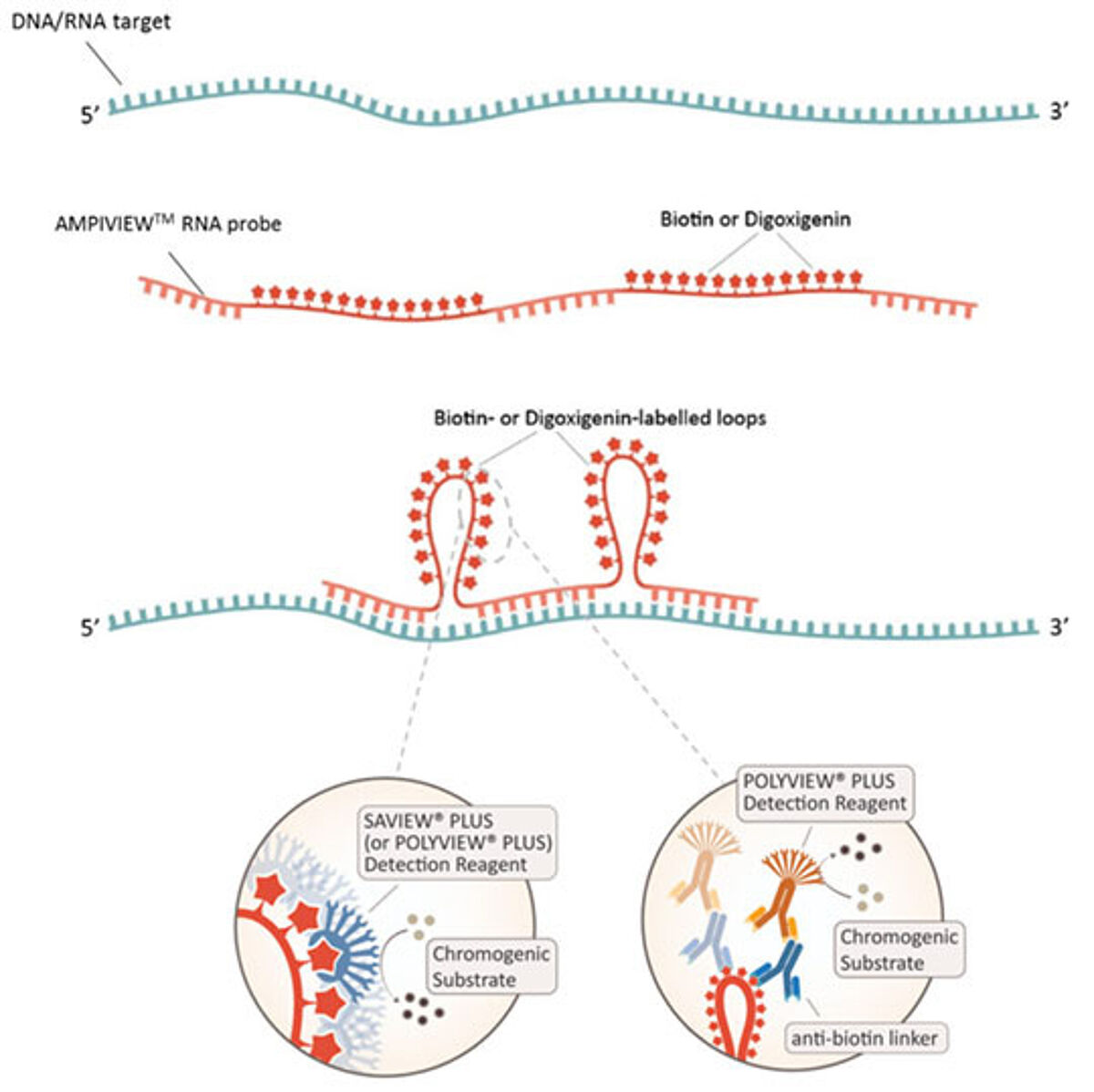
Enzo Life Sciences, considered a pioneer in labeling and detection since the development of the first non-radioactive human papillomavirus (HPV) probes for ISH in 1986, is today proud to give a further contribution in the field with the introduction of the AMPIVIEW™ RNA probes, based on the patented LoopRNA technology. As represented in figure 3, in a LoopRNA ISH probe (LRP), the regions pairing with the target sequence are interspersed with multiple RNA spacer segments, which are not complementary to the target. The spacers are designed so that most of their bases can be labeled with either biotin or digoxigenin. When such a probe is hybridized to its target sequence, the labeled spacer segments loop out and can then be recognized by the reporter system (5). The presence of multiple molecules of biotin or digoxigenin, easily accessible for the reporters thanks to the loops, allow for a strong amplification of the signal, thus assuring an excellent sensitivity. Since the hybridization does not need to undergo multiple amplification steps, the protocol is much faster and easier compared to the bDNA system described above. In addition, the AMPIVIEW™ probes do not require any specific equipment, and can be used in manual or automated procedures.
Concerning the detection, pretty much as for a standard ISH, it can be direct or indirect. When the probe is labeled with biotin, a streptavidin conjugated with a reporter enzyme can be directly used (direct or one-step protocol). Enzo’s SAVIEW® PLUS AP or HRP Reagent is a ready-to-use streptavidin-based nanopolymer reagent, ensuring consistent and reproducible results. In the indirect (or two-step) protocol, anti-biotin or anti-digoxigenin linker antibodies can be combined with ready-to-use nanopolymer POLYVIEW® PLUS AP or HRP detection reagents, producing high intensity color development, with sharp and crisp staining. Stunning results can then be achieved when combining SAVIEW® PLUS or POLYVIEW® PLUS reagents with Enzo’s unrivaled selection of HIGHDEF® chromogens (9). For all these reasons, AMPIVIEW™ RNA probes represent a cost-effective solution and easily adaptable into any laboratory.
Faithful to our historical background, the first AMPIVIEW™ RNA probes generated were to target HPV. In a recent collaboration between Enzo and two Italian universities, Dr. La Rocca et al. demonstrated that ISH probes based on the LoopRNA technology allow the detection of HPV DNA and RNA even down to a single copy of genome-integrated HPV. These results are compared with another commercially available double-Z probe, demonstrating the equivalent or superior performance of the AMPIVIEW™ RNA probes (5). The article also highlights another relevant feature of LoopRNAs, which is the possibility to synthetize probes corresponding to both sense (complimentary to the viral genomic DNA) and antisense sequence (complimentary to genomic DNA and RNA transcript) in order to distinguish between genomic DNA and RNA being actively transcribed by the virus.
It is also important to mention here that AMPIVIEW™ RNA probes can be designed for practically any gene, in any genome. For instance, during the COVID-19 pandemic, Dr. Nuovo from Ohio University used an AMPIVIEW™ probe to detect SARS-CoV-2 RNA expression in different tissues from COVID-19 patients, comparing the presence of viral genome with the presence of the four characteristic viral protein (envelope, spike, membrane, and nucleocapsid) (8). Dr. Nuovo has also been a valuable guest speaker in webinar series to illustrate these results. You can re-watch its brilliant talk HERE!
Products
- AMPIVIEW™ RNA probes
Do you have questions regarding our AMPIVIEW™ RNA probes, powered by Enzo’s LoopRNA™ technologies? If you are looking for a probe for a specific target and would like to test the AMPIVIEW solution, do not hesitate to contact us. We will be happy to assist!
More about AMPIVIEW
- Stellaris® RNA probes
Stellaris® RNA FISH is a RNA visualization method that allows simultaneous detection, localization, and quantification of individual mRNA molecules at the cellular level in fixed samples.
Suppliers

Enzo Life Sciences
Enzo Life Sciences is a leading life sciences and biotechnology company offering assay kits, imaging probes, antibodies, proteins and small molecules.

LGC Biosearch Technologies
LGC Biosearch Technologies provides the well-known Stellaris® RNA FISH probes, and other products and services for genomic analysis.
References
- Gall JG & Pardue ML. Formation and detection of RNA-DNA hybrid molecules in cytological preparations. PNAS (1969). PMID: 4895535.
- John HA et al. RNA-DNA hybrids at the cytological level. Nature (1969). PMID: 5799530.
- FarrellJr RE. Nucleic Acid Probe Technology. RNA Methodologies, Ch 12. (2020). Full Text.
- Wang F et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn (2012). PMID: 22166544.
- La Rocca G et al. Recent improvements in in situ hybridization for the detection of HPV infections in clinical samples. WCRJ (2020). Full Text.
- Player AN et al. Single-copy gene detection using branched DNA (bDNA) in situ hybridization. J Histochem Cytochem. (2001). PMID: 11304798.
- Atout S et al. Evaluation of the Suitability of RNAscope as a Technique to Measure Gene Expression in Clinical Diagnostics: A Systematic Review. Mol Diagn Ther (2022). PMID: 34957535.
- Nuovo GJ et al. A Standardization Protocol for the In Situ Detection of SARS-CoV2 RNA and Proteins. Appl Immunohistochem Mol Morphol. (2022). PMID: 35175238.
- Nuovo GJ. False-positive results in a diagnostic immunohistochemistry are related to horseradish peroxidase conjugates in commercially available assays. Annals of Diagnostic Pathology (2016). PMID 27806847.